|
|
| (21 intermediate revisions by 2 users not shown) |
| Line 1: |
Line 1: |
| | + | <center>[[CTK|CTK home]] | [[Standard/BrdU-CLIP data analysis using CTK|Standard/BrdU-CLIP]] | [[iCLIP data analysis using CTK|iCLIP]] | [[eCLIP data analysis using CTK|eCLIP]] | [[PARCLIP data analysis using CTK|PAR-CLIP]] | [[CTK_usage|CTK usage]] | [[CTK_FAQ|FAQ]]</center> |
| | + | |
| | + | __TOC__ |
| | + | |
| | =Introduction= | | =Introduction= |
| | | | |
| | [[File:CTK_Pipeline_Overview.png|500px|center]] | | [[File:CTK_Pipeline_Overview.png|500px|center]] |
| | | | |
| − | <center>'''[[CTK_usage|CTK usage]] | [[CTK_FAQ|FAQ]]'''</center>
| |
| | | | |
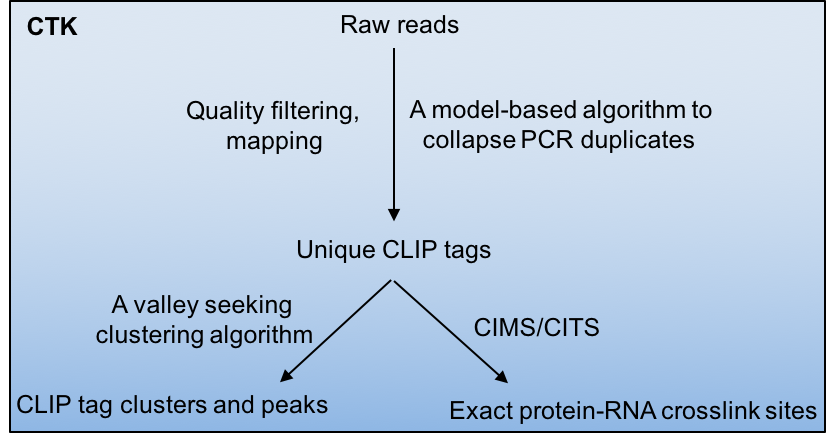
| − | Crosslinking and immunoprecipitation paired with highthroughput sequencing (HITS-CLIP or CLIP-Seq) has now been widely used to map protein-RNA interactions on a genome-wide scale. The CLIP Tool Kit (CTK) is a software package that provides a set of tools for analysis of CLIP data starting from the raw reads generated by the sequencer. It includes pipelines to filter and map reads, collapse PCR duplicates to obtain unique CLIP tags, define CLIP tag clusters and call peaks, and define the exact protein-RNA crosslink sites by CIMS and CITS analysis. This software package is an expansion of our previous CIMS package. | + | Crosslinking and immunoprecipitation followed by highthroughput sequencing (HITS-CLIP or CLIP-Seq) has now been widely used to map protein-RNA interactions on a genome-wide scale. The CLIP Tool Kit (CTK) is a software package that provides a set of tools for analysis of CLIP data starting from the raw reads generated by the sequencer. It includes pipelines to filter and map reads, collapse PCR duplicates to obtain unique CLIP tags, define CLIP tag clusters and call peaks, and define the exact protein-RNA crosslink sites by CIMS and CITS analysis. This software package is an expanded version of our previous CIMS package. |
| | | | |
| − | Crosslinking induced mutation site (CIMS) analysis is a computational method for CLIP-Seq data analysis to determine exact protein-RNA crosslink sites and thereby map protein-RNA interactions at single-nucleotide resolution. This method is based on the observation that UV cross linked amino-acid-RNA adducts introduce reverse transcription errors in cDNAs at certain frequencies, which are captured by sequencing and subsequent comparison of CLIP tags with a reference genome. | + | Crosslinking induced mutation site (CIMS) and cross linking induced truncation site (CITS) analyses are computational methods for CLIP data analysis to determine the exact protein-RNA crosslink sites and thereby map protein-RNA interactions at single-nucleotide resolution. These methods are based on the observation that UV crosslinked amino-acid-RNA adducts can introduce reverse transcription errors, including mutations and premature in cDNAs at a certain frequency, which are captured by sequencing and subsequent comparison of CLIP tags with a reference genome. |
| | | | |
| | | | |
| Line 16: |
Line 19: |
| | | | |
| | | | |
| − | More details of the biochemical and computational aspects of CLIP-Seq can be found in the following references: | + | More details of the biochemical and computational aspects of CLIP can be found in the following references: |
| | | | |
| | <pre> | | <pre> |
| Line 27: |
Line 30: |
| | | | |
| | <pre> | | <pre> |
| − | Weyn-Vanhentenryck,S.,M.*, Mele,A.*, Yan,Q.*, Sun,S., Farny,N., Zhang,Z., Xue,C., Herre,M., Silver,P.A., Zhang,M.Q., Krainer,A.R., Darnell,R.B. †, Zhang,C. † 2014. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 10.1016/j.celrep.2014.02.005. | + | Weyn-Vanhentenryck,S.,M.*, Mele,A.*, Yan,Q.*, Sun,S., Farny,N., Zhang,Z., Xue,C., Herre,M., Silver,P.A., Zhang,M.Q., Krainer,A.R., Darnell,R.B. †, Zhang,C. † 2014. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 6:1139-1152. |
| | </pre> | | </pre> |
| | + | |
| | + | =User group= |
| | + | |
| | + | For questions/answers, please visit our user group: https://groups.google.com/forum/#!forum/ctk-user-group |
| | | | |
| | =Versions= | | =Versions= |
| − | *v1.0.7 ( 01-16-2017) current | + | *v1.1.3 (12/2018) current |
| | + | **minor bug fix |
| | + | *v1.1.2 (08/02/2018) |
| | + | **update in hg38 annotation files |
| | + | *v1.1.1 (07/20/2018) |
| | + | ** bug fix related to the use of dm6 annotation files |
| | + | *v1.1.0 ( 07/14/2018 ) |
| | + | ** included support for dm6. |
| | + | *v1.0.9 ( 06/12/2018 ) |
| | + | ** minor bug fix |
| | + | *v1.0.8 ( 05/24/2018 ) |
| | + | **improved selection of unique CLIP tags. |
| | + | **improved support for CIMS anlaysis of particular types of substitutions (e.g., T-C for PAR-CLIP). |
| | + | **a wrapper CITS.pl is included to simplify CITS anlaysis. |
| | + | **included additional annotations files. |
| | + | **included support for hg38. |
| | + | **minor bug fixes |
| | + | **improved/expanded documentation and tutorials |
| | + | **Various improvement and bug fixes to improve efficiency and robustness |
| | + | *v1.0.7 ( 01-16-2017 ) |
| | **fix mac-specific crash | | **fix mac-specific crash |
| − | *v1.0.6 ( 01-04-2017) | + | *v1.0.6 ( 01-04-2017 ) |
| | **minor bug fix | | **minor bug fix |
| − | *v1.0.5 (11-10-2016) | + | *v1.0.5 ( 11-10-2016 ) |
| | **fixed path to annotation files | | **fixed path to annotation files |
| | **have default path to gene bed file for tag2peak.pl | | **have default path to gene bed file for tag2peak.pl |
| − | *v1.0.4 ( 10-05-2016) | + | *v1.0.4 ( 10-05-2016 ) |
| | **minor fixes | | **minor fixes |
| | *v1.0.3 ( 08-08-2016 ) | | *v1.0.3 ( 08-08-2016 ) |
| Line 47: |
Line 73: |
| | =Download= | | =Download= |
| | | | |
| − | ==Source code==
| + | *czplib (perl): a perl library with various functions for genomic/bioinformatic analysis. [https://github.com/chaolinzhanglab/czplib Download from github] |
| − | | + | *CTK (perl): the core algorithm. [https://github.com/chaolinzhanglab/ctk Download from github] |
| − | *czplib (perl): a perl library with various functions for genomic/bioinformatic analysis. [https://github.com/chaolinzhanglab/czplib/tree/v1.0.7 download from github] | + | |
| − | *CTK (perl): the core algorithm. [https://github.com/chaolinzhanglab/ctk/ download from github] | + | |
| − | | + | |
| − | ==Sample dataset==
| + | |
| − | | + | |
| − | '''Raw FASTQ files:'''
| + | |
| − | | + | |
| − | In this documentation, we will use the sample mouse brain Rbfox1-3 CLIP data (Weyn-Vanhentenryck et al. Cell Reports 2014) as a guide. Two protocols (standard CLIP and BrdU-CLIP) were used to generate this dataset.
| + | |
| − | | + | |
| − | The raw sequence files from Illumina sequencing can be downloaded from SRA (http://www.ncbi.nlm.nih.gov/sra/?term=SRP035321), and we assume FASTQ files have been generated using the sra toolkit (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?cmd=show&f=software&m=software&s=software) and the following files Fox1_1.fastq.gz, Fox1_2.fastq.gz, Fox1_3.fastq.gz, Fox2_1.fastq.gz, Fox2_2.fastq.gz, Fox2_3.fastq.gz, Fox2_4.fastq.gz, Fox3_1.fastq.gz, Fox3_2.fastq.gz, Fox3_3.fastq.gz, Fox3_4.fastq.gz, Fox3_5.fastq.gz can be moved to the directory called ''fastq'' under the ''test'' folder in home directory (i.e., ~/test is our working directory). Fox1_1.fastq.gz, Fox2_1.fastq.gz, and Fox3_1.fastq.gz were generated from the Standard CLIP protocol, and the rest were generated from the BrdU CLIP protocol.
| + | |
| − | | + | |
| − | | + | |
| − | '''Download sample output files of major steps here:'''
| + | |
| − | | + | |
| − | *[http://zhanglab.c2b2.columbia.edu/data/CTK/preprocess.tar.gz Fully preprocessed FASTQ files] : output files generated for use right before mapping. The samples have been quality filtered, trimmed of 3' linker sequences, had exact duplicates collapsed, and barcodes stripped.
| + | |
| − | | + | |
| − | *[http://zhanglab.c2b2.columbia.edu/data/CTK/unique.tar.gz Unique CLIP tag and mutation files] : output files generated after mapping and collapsing PCR duplicates.
| + | |
| − | | + | |
| − | *[http://zhanglab.c2b2.columbia.edu/data/CTK/peak_calling.tar.gz Peak calling files] : pooled Rbfox unique tag bed file as input + output files after peak calling.
| + | |
| − | | + | |
| − | *[http://zhanglab.c2b2.columbia.edu/data/CTK/CIMS.tar.gz CIMS output files] : output files after CIMS analysis.
| + | |
| − | | + | |
| − | *[http://zhanglab.c2b2.columbia.edu/data/CTK/CITS.tar.gz CITS output files] : output files after CITS analysis.
| + | |
| | | | |
| | =Prerequisites= | | =Prerequisites= |
| | | | |
| − | This software is implemented in perl. It also relies on several standard linux/unix tools such as grep, cat, sort, etc. We have tested the software on RedHat Linux (Linux 2.6.32-504.8.1.el6.x86_64), although it is expected to work on most unix-like systems, including Mac OS X. In addition, several software packages are required by the pipeline for sequence preprocessing and alignment (the version number in our test is also indicated). | + | This software is implemented in perl. It also relies on several standard linux/unix tools such as grep, cat, sort, etc. We have tested the software on Cent OS, although it is expected to work on most unix-like systems, including Mac OS X. In addition, several software packages are required by the pipeline for sequence preprocessing and alignment (the version number in our test is also indicated). |
| | | | |
| | *FASTX Tool-Kit Version 0.0.13: http://hannonlab.cshl.edu/fastx_toolkit/download.html | | *FASTX Tool-Kit Version 0.0.13: http://hannonlab.cshl.edu/fastx_toolkit/download.html |
| | + | *cutadapt Version 1.14: https://pypi.python.org/pypi/cutadapt/ (an alternative to FASTX Tool-Kit) |
| | *Burrows Wheeler Aligner (BWA) Version 0.7.12: http://bio-bwa.sourceforge.net/ | | *Burrows Wheeler Aligner (BWA) Version 0.7.12: http://bio-bwa.sourceforge.net/ |
| | *Samtools Version 1.3.1: http://samtools.sourceforge.net | | *Samtools Version 1.3.1: http://samtools.sourceforge.net |
| Line 85: |
Line 89: |
| | =Installation= | | =Installation= |
| | | | |
| − | * Download and install software packages described in prerequisites.
| + | ==Through anaconda== |
| − | * Download the czplib perl library files (refer back to '''Download''' section above)
| + | |
| − | * Decompress and move to whatever directory you like (as an example, we use /usr/local/lib/)
| + | |
| − | * Replace "x.tgz" below with the version of the package you downloaded
| + | |
| − | <pre>
| + | |
| − | $unzip czplib-1.0.x.zip
| + | |
| − | $mv czplib-1.0.x /usr/local/lib/czplib
| + | |
| − | </pre>
| + | |
| | | | |
| − | Add the library path to the environment variable, so perl can find it.
| + | Below are the installation instructions for the perl packages CTK and CZPLIB through Anaconda. |
| − | <pre>
| + | |
| − | export PERL5LIB=/usr/local/lib/czplib
| + | |
| − | </pre>
| + | |
| | | | |
| − | * Download CTK code and likewise decompress and move to whatever directory you like (as an example, we use /usr/local/) | + | * Setup the working environment 'ctk' and install all the packages by running the commands below: |
| | | | |
| | <pre> | | <pre> |
| − | $unzip ctk-1.0.x.zip
| + | myenv='ctk' |
| − | $mv ctk-1.0.x /usr/local/CTK | + | conda create --yes --name $myenv |
| − | </pre>
| + | |
| | | | |
| − | Add the dir to your $PATH environment variable if you would like.
| + | conda activate $myenv |
| | | | |
| − | Finally, some of the scripts will use a cache directory, which is under the working directory by default. One can specify another folder for cache using environment variable.
| + | conda config --env --append channels conda-forge |
| | + | conda config --env --append channels bioconda |
| | | | |
| − | <pre>
| + | conda install --yes -c chaolinzhanglab ctk |
| − | | + | |
| − | #e.g., add the following lines in .bash_profile
| + | |
| − | CACHEHOME=$HOME/cache
| + | |
| − | export CACHEHOME
| + | |
| | </pre> | | </pre> |
| | | | |
| − | =Read preprocessing=
| + | If you would like to install a specific platform version e.g. 'noarch', then use the below command: |
| − | | + | |
| − | Compared to our previous software package, this new pipeline, in all preprocessing steps prior to read alignment, operates on FASTQ files to take advantage of sequence quality scores.
| + | |
| − | | + | |
| − | ==Sample dataset==
| + | |
| − | In our current experimental design (BrdU-CLIP), several CLIP libraries with different indexes are typically pooled together to be sequenced in one lane.
| + | |
| − | | + | |
| − | The sample dataset we use as a guide (mouse brain Rbfox1-3 CLIP) was generated using the standard CLIP or BrdU-CLIP protocols (see the table below; see the Download section if you have not downloaded them). We assume FASTQ files have been generated using the sra toolkit (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?cmd=show&f=software&m=software&s=software) and the following files Fox1_1.fastq.gz, Fox1_2.fastq.gz, Fox1_3.fastq.gz, Fox2_1.fastq.gz, Fox2_2.fastq.gz, Fox2_3.fastq.gz, Fox2_4.fastq.gz, Fox3_1.fastq.gz, Fox3_2.fastq.gz, Fox3_3.fastq.gz, Fox3_4.fastq.gz, Fox3_5.fastq.gz can be moved to the directory called ''fastq'' under the ''test'' folder in home directory (i.e., ~/test is our working directory). Fox1_1.fastq.gz, Fox2_1.fastq.gz, and Fox3_1.fastq.gz were generated from the Standard CLIP protocol, and the rest were generated from the BrdU CLIP protocol.
| + | |
| − | | + | |
| − | Reads from the standard CLIP protocol begin with a 5nt random barcode, followed by the actual cDNA tags of variable lengths, and possibly 3' adaptor sequences.
| + | |
| − | | + | |
| − | Reads from the BrdU-CLIP protocol begin with a 14 nucleotides that are sample index and random barcode (4 nt sample index+ a fixed base (either G or C)+ an 8-nt random barcode + a G), followed by the actual cDNA tags of variable lengths, and possibly 3' adaptor sequences.
| + | |
| − | | + | |
| − | | + | |
| − | The number of reads in each file, after each step of processing, is summarized in Table 1 below.
| + | |
| − | {| class="wikitable"
| + | |
| − | |+ Table 1: Summary of read numbers analyzed by CTK.
| + | |
| − | ! Protein
| + | |
| − | ! Sample
| + | |
| − | ! CLIP protocol
| + | |
| − | ! # of raw reads
| + | |
| − | ! # of filtered reads
| + | |
| − | ! # of trimmed reads
| + | |
| − | ! # of collapsed reads
| + | |
| − | ! # of mapped reads
| + | |
| − | ! # of unique tags
| + | |
| − | |-
| + | |
| − | | Fox1
| + | |
| − | | Fox1_1
| + | |
| − | | Standard
| + | |
| − | | 128,502,865
| + | |
| − | | 124,979,444
| + | |
| − | | 113,958,244
| + | |
| − | | 4,953,805
| + | |
| − | | 3,713,799
| + | |
| − | | 357,094
| + | |
| − | |-
| + | |
| − | | Fox1
| + | |
| − | | Fox1_2
| + | |
| − | | BrdU
| + | |
| − | | 42,373,740
| + | |
| − | | 39,121,986
| + | |
| − | | 35,683,047
| + | |
| − | | 1,465,789
| + | |
| − | | 1,098,974
| + | |
| − | | 94,757
| + | |
| − | |-
| + | |
| − | | Fox1
| + | |
| − | | Fox1_3
| + | |
| − | | BrdU
| + | |
| − | | 36,103,020
| + | |
| − | | 32,500,181
| + | |
| − | | 29,682,907
| + | |
| − | | 2,112,635
| + | |
| − | | 1,468,725
| + | |
| − | | 161,487
| + | |
| − | |-
| + | |
| − | | Fox1
| + | |
| − | | Fox1_4
| + | |
| − | | BrdU
| + | |
| − | | 64,528,510
| + | |
| − | | 52,712,783
| + | |
| − | | 48,292,964
| + | |
| − | | 7,025,945
| + | |
| − | | 5,262,144
| + | |
| − | | 1,194,617
| + | |
| − | |-
| + | |
| − | | Fox2
| + | |
| − | | Fox2_1
| + | |
| − | | Standard
| + | |
| − | | 135,171,343
| + | |
| − | | 131,229,217
| + | |
| − | | 121,238,246
| + | |
| − | | 5,406,496
| + | |
| − | | 3,716,401
| + | |
| − | | 295,180
| + | |
| − | |-
| + | |
| − | | Fox2
| + | |
| − | | Fox2_2
| + | |
| − | | BrdU
| + | |
| − | | 42,744,272
| + | |
| − | | 39,351,367
| + | |
| − | | 35,005,375
| + | |
| − | | 1,314,878
| + | |
| − | | 968,115
| + | |
| − | | 83,358
| + | |
| − | |-
| + | |
| − | | Fox2
| + | |
| − | | Fox2_3
| + | |
| − | | BrdU
| + | |
| − | | 31,512,102
| + | |
| − | | 28,530,199
| + | |
| − | | 25,877,412
| + | |
| − | | 2,356,210
| + | |
| − | | 1,581,335
| + | |
| − | | 337,811
| + | |
| − | |-
| + | |
| − | | Fox2
| + | |
| − | | Fox2_4
| + | |
| − | | BrdU
| + | |
| − | | 46,383,375
| + | |
| − | | 40,317,796
| + | |
| − | | 29,418,988
| + | |
| − | | 2,761,500
| + | |
| − | | 1,803,408
| + | |
| − | | 165,645
| + | |
| − | |-
| + | |
| − | | Fox3
| + | |
| − | | Fox3_1
| + | |
| − | | Standard
| + | |
| − | | 181,578,804
| + | |
| − | | 173,884,893
| + | |
| − | | 162,979,630
| + | |
| − | | 10,405,177
| + | |
| − | | 7,742,410
| + | |
| − | | 877,256
| + | |
| − | |-
| + | |
| − | | Fox3
| + | |
| − | | Fox3_2
| + | |
| − | | BrdU
| + | |
| − | | 38,828,880
| + | |
| − | | 35,983,470
| + | |
| − | | 27,171,512
| + | |
| − | | 1,185,180
| + | |
| − | | 857,925
| + | |
| − | | 143,522
| + | |
| − | |-
| + | |
| − | | Fox3
| + | |
| − | | Fox3_3
| + | |
| − | | BrdU
| + | |
| − | | 31,774,690
| + | |
| − | | 29,095,065
| + | |
| − | | 26,332,663
| + | |
| − | | 2,295,078
| + | |
| − | | 1,669,958
| + | |
| − | | 399,878
| + | |
| − | |-
| + | |
| − | | Fox3
| + | |
| − | | Fox3_4
| + | |
| − | | BrdU
| + | |
| − | | 32,658,459
| + | |
| − | | 29,720,851
| + | |
| − | | 21,737,823
| + | |
| − | | 2,017,979
| + | |
| − | | 1,479,115
| + | |
| − | | 413,847
| + | |
| − | |-
| + | |
| − | | Fox3
| + | |
| − | | Fox3_5
| + | |
| − | | BrdU
| + | |
| − | | 58,175,879
| + | |
| − | | 47,324,888
| + | |
| − | | 42,941,116
| + | |
| − | | 6,200,102
| + | |
| − | | 4,551,275
| + | |
| − | | 1,076,782
| + | |
| − | |-
| + | |
| − | | '''Total'''
| + | |
| − | |
| + | |
| − | |
| + | |
| − | | '''870,706,469'''
| + | |
| − | |
| + | |
| − | |
| + | |
| − | |
| + | |
| − | |
| + | |
| − | | '''5,601,234'''
| + | |
| − | |}
| + | |
| − | | + | |
| − | ==Demultiplexing samples ==
| + | |
| − | | + | |
| − | The files downloaded from SRA have already be demultiplexed. Therefore the next step has already been performed. It is included for illustration purposes for the user who would like to start from a FASTQ file consisting of multiple libraries.
| + | |
| − | | + | |
| | <pre> | | <pre> |
| − | cd ~/test/fastq
| + | conda install --yes -c chaolinzhanglab/noarch ctk |
| − | | + | |
| | </pre> | | </pre> |
| | | | |
| − | Assuming that we have six samples with indexes GTCA, GCATG, ACTG, AGCT, GCATC, TCGA, the following script will extract reads for each library using a simple loop.
| + | For ease of installation, we recommend the above commands to be copied to a script setup_ctk.sh. This will also make it easy to include the required steps (See '''Note''' below) for restoring the PATH variable if we want to. |
| | | | |
| | + | Assuming the conda base environment is already setup and activated, run the setup_ctk.sh script from the terminal. |
| | <pre> | | <pre> |
| − | #!/bin/bash
| + | (base)...$source setup_ctk.sh |
| − | #For illustration
| + | |
| − | | + | |
| − | index=(GTCA GCATG ACTG AGCT GCATC TCGA);
| + | |
| − | samples=(sample1 sample2 sample3 sample4 sample5 sample6);
| + | |
| − | | + | |
| − | for ((i=0;i<=5;i=i+1)); do
| + | |
| − | idx=${index[$i]}
| + | |
| − | name=${samples[$i]}
| + | |
| − | perl /usr/local/CTK/fastq_filter.pl -v -if sanger -index 0:$idx -of fastq raw.fastq.gz - | gzip -c > $name.fastq.gz
| + | |
| − | done
| + | |
| | </pre> | | </pre> |
| | | | |
| − | Alternatively, we can parallelize the process by submitting an array of jobs to SGE.
| |
| | | | |
| − | <pre>
| + | Now we can proceed to our working directory for performing the CTK analysis. |
| | | | |
| − | #!/bin/bash
| |
| − | #$ -t 1-6 -m a -cwd -N CLIP
| |
| − | #For illustration
| |
| | | | |
| − | index=(GTCA GCATG ACTG AGCT GCATC TCGA);
| + | '''Acknowledgment Note:''' |
| − | samples=(sample1 sample2 sample3 sample4 sample5 sample6);
| + | There is a R wrapper for CTK, called CLIPflexR which can call some other external libraries within R. |
| | | | |
| | + | For details please visit the CLIPflexR webpage: |
| | | | |
| − | idx=${index[$SGE_TASK_ID-1]}
| + | https://kathrynrozengagnon.github.io/CLIPflexR/index.html |
| − | name=${samples[$SGE_TASK_ID-1]}
| + | |
| − | perl /usr/local/CTK/fastq_filter.pl -v -if sanger -index 0:$idx -of fastq raw.fastq.gz - | gzip -c > $name.fastq.gz
| + | |
| | | | |
| − | </pre>
| |
| | | | |
| − | The script /usr/local/CTK/fastq_filter.pl will extract reads with indicated index sequence (e.g., GTCA) starting from position 0 (specified by the -index option) in the read.
| |
| | | | |
| − | *Usage and additional explanation of [[CTK_usage#fastq_filter.pl|fastq_filter.pl]]
| + | '''Note:''' |
| − | | + | If we need to reset the PATH variable when we do 'conda deactivate' to go to the base environment from the working environment 'ctk', we suggest to include the following steps in the setup_ctk.sh. |
| − | '''Note 1''': This script can also be used to demultiplex samples from other CLIP protocols as long as the position relative to the read start is fixed. | + | This is a crude way but serves the purpose. |
| − | | + | |
| − | '''Note 2''': To get usage information, just run the script without any parameters (the same for the other scripts in the package). | + | |
| − | | + | |
| − | ==Read quality filtering==
| + | |
| | | | |
| | + | 1. At the beginning of the script setup_ctk.sh, include the following: |
| | <pre> | | <pre> |
| − | cd ~/test
| + | export CONDA_PATH_RESET=/usr/local/bin/:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:${CONDA_PREFIX}/bin:${CONDA_PREFIX}/condabin |
| − | mkdir filtering
| + | |
| − | cd filtering
| + | |
| | </pre> | | </pre> |
| | | | |
| − | The same fastq_filter.pl script will extract reads passing quality filters for each library (of standard CLIP-seq data) using a simple loop.
| + | 2. Just before the line "conda activate $myenv", include the following: |
| − | | + | |
| | <pre> | | <pre> |
| − | #!/bin/bash
| + | echo "CONDA_PATH_RESET=$CONDA_PATH_RESET" > "${CONDA_PREFIX}/envs/${myenv}/etc/conda/activate.d/${myenv}_env_activate.sh" |
| | + | echo "export CONDA_PATH_RESET" >> "${CONDA_PREFIX}/envs/${myenv}/etc/conda/activate.d/${myenv}_env_activate.sh" |
| | | | |
| − | for f in Fox1_1 Fox2_1 Fox3_1
| + | chmod +x "${CONDA_PREFIX}/envs/${myenv}/etc/conda/activate.d/${myenv}_env_activate.sh" |
| − | do
| + | |
| − | perl /usr/local/CTK/fastq_filter.pl -v -if sanger -f mean:0-29:20 -of fastq ../fastq/$f.fastq.gz $f.fastq
| + | |
| − | done
| + | |
| | </pre> | | </pre> |
| | | | |
| − | | + | 3. Just after the line "conda activate $myenv", include the following: |
| − | This is the same thing, but using a Sun Grid Engine to execute this UNIX job on a remote machine.
| + | |
| | <pre> | | <pre> |
| − | #!/bin/bash
| + | echo "PATH=\$CONDA_PATH_RESET" > "${CONDA_PREFIX}/etc/conda/deactivate.d/${myenv}_env_deactivate.sh" |
| − | #$ -t 1-3 -m a -cwd -N CLIP
| + | echo "export PATH" >> "${CONDA_PREFIX}/etc/conda/deactivate.d/${myenv}_env_deactivate.sh" |
| − | | + | |
| − | files=(Fox1_1 Fox2_1 Fox3_1);
| + | |
| − | f=${files[$SGE_TASK_ID-1]}
| + | |
| − | | + | |
| − | perl /usr/local/CTK/fastq_filter.pl -v -if sanger -f mean:0-29:20 -of fastq ../fastq/$f.fastq.gz $f.fastq
| + | |
| | | | |
| | + | chmod +x "${CONDA_PREFIX}/etc/conda/deactivate.d/${myenv}_env_deactivate.sh" |
| | </pre> | | </pre> |
| | | | |
| − | This script filters raw reads based on quality scores. -f mean:0-29-20 specifies a mean score of 20 or above in the first 30 bases (i.e., positions 0-29 for Standard CLIP), which includes 5 positions with sample indexes and the random barcode, followed by 25 positions with the actual CLIP tag.
| + | These steps will write the required activate.d and deactivate.d scripts to reset the $PATH variable |
| | | | |
| − | The filtering should be run again on the BrdU CLIP (or iCLIP) samples slightly differently.
| |
| | | | |
| − | <pre>
| |
| − | #!/bin/bash
| |
| − | #$ -t 1-10 -m a -cwd -N CLIP
| |
| − |
| |
| − | files=(Fox1_2 Fox1_3 Fox1_4 Fox2_2 Fox2_3 Fox2_4 Fox3_2 Fox3_3 Fox3_4 Fox3_5);
| |
| − | f=${files[$SGE_TASK_ID-1]}
| |
| − |
| |
| − | perl /usr/local/CTK/fastq_filter.pl -v -if sanger -f mean:0-38:20 -of fastq ../fastq/$f.fastq.gz $f.fastq
| |
| − | </pre>
| |
| − |
| |
| − | -f mean:0-38-20 specifies a mean score of 20 or above in the first 39 bases (i.e., positions 0-38 for BrdU CLIP), which includes 14 positions with sample indexes and the random barcode, followed by 25 positions with the actual CLIP tag.
| |
| − |
| |
| − | The reason we filter as such is because low quality reads can introduce mapping errors and background. They will inflate the number of unique tags after removal of PCR duplicates.
| |
| − |
| |
| − | '''Note 1:''' The reason we used the first four positions of the index for demultiplexing is due to historical confusion about the identity of the index sequences. It really should be 5nt, but it does not make a substantial difference.
| |
| − |
| |
| − | '''Note 2:''' As a good practice, always check the number of reads in the raw files and compare the number of reads left after filtering to confirm if the numbers are what you would expect. This can be done by running the command:
| |
| | | | |
| | + | ==Manual Installation== |
| | + | * Download and install software packages described in prerequisites. |
| | + | * Download the czplib perl library files (refer back to '''Download''' section above) |
| | + | * Decompress and move to whatever directory you like (as an example, we use /usr/local/lib/) |
| | + | * Replace "x.tgz" below with the version of the package you downloaded |
| | <pre> | | <pre> |
| − | #for raw reads
| + | $unzip czplib-1.0.x.zip |
| − | for f in `ls ../fastq/*.fastq.gz`; do
| + | $mv czplib-1.0.x /usr/local/lib/czplib |
| − | zcat $f | wc -l
| + | |
| − | done
| + | |
| − | | + | |
| − | #for filtered reads
| + | |
| − | wc -l *.fastq
| + | |
| | </pre> | | </pre> |
| | | | |
| − | The '''wc''' command will give the number of lines (divide by 4 to get the actual read number).
| + | Add the library path to the environment variable, so perl can find it. |
| − | | + | |
| − | '''Note 3:''' Demultiplexing and quality filtering can be combined into a single step by specifying -index and -f at the same time.
| + | |
| − | | + | |
| | <pre> | | <pre> |
| − | perl /usr/local/CTK/fastq_filter.pl -v -if sanger -index 0:GTCA -f mean:0-38:20 -of fastq ../fastq/in.fastq.gz - | gzip -c > out.fastq.gz
| + | export PERL5LIB=/usr/local/lib/czplib |
| | </pre> | | </pre> |
| | | | |
| − | ==Trimming of 3' linker sequences==
| + | * Download CTK code and likewise decompress and move to whatever directory you like (as an example, we use /usr/local/) |
| − | | + | |
| − | For long reads that are common now, collapsing before trimming is not very helpful. Therefore, we trim the 3' linker first (which can vary depending on the specific CLIP protocol performed).
| + | |
| − | | + | |
| − | For each sample, run the following command using tools provided by the Fastx Toolkit.
| + | |
| | | | |
| | <pre> | | <pre> |
| − | #!/bin/bash
| + | $unzip ctk-1.0.x.zip |
| − | #$ -t 1-3 -m a -cwd -N CLIP
| + | $mv ctk-1.0.x /usr/local/CTK |
| − | #-l "mem=2G,time=192::"
| + | |
| − | | + | |
| − | files=(Fox1_1 Fox2_1 Fox3_1);
| + | |
| − | f=${files[$SGE_TASK_ID-1]}
| + | |
| − | | + | |
| − | fastx_clipper -a GTGTCAGTCACTTCCAGCGG -l 20 -n -i $f.fastq | fastq_quality_trimmer -t 5 -l 20 -o $f.trim.fastq #note that the 3' linker will vary for other CLIP protocol variations.
| + | |
| | </pre> | | </pre> |
| | | | |
| − | This command will remove the 3' linker and/or also remove extremely low quality bases (score < 5).
| + | Add the dir to your $PATH environment variable if you would like. |
| | | | |
| − | You can do the same for the BrdU CLIP files:
| + | Finally, some of the scripts will use a cache directory, which is under the working directory by default. One can specify another folder for cache using environment variable (recommended). |
| | | | |
| | <pre> | | <pre> |
| − | #!/bin/bash
| |
| − | #$ -t 1-10 -m a -cwd -N CLIP
| |
| − | #-l "mem=2G,time=192::"
| |
| − | files=(Fox1_2 Fox1_3 Fox1_4 Fox2_2 Fox2_3 Fox2_4 Fox3_2 Fox3_3 Fox3_4 Fox3_5);
| |
| − | f=${files[$SGE_TASK_ID-1]}
| |
| − |
| |
| − | fastx_clipper -a TCGTATGCCGTCTTCTGCTTG -l 29 -n -i $f.fastq | fastq_quality_trimmer -t 5 -l 29 -o $f.trim.fastq
| |
| | | | |
| | + | #e.g., add the following lines in .bash_profile |
| | + | CACHEHOME=$HOME/cache |
| | + | export CACHEHOME |
| | </pre> | | </pre> |
| | | | |
| − | For fastx_clipper usage information:
| + | =Indexing reference genome = |
| − | <pre>
| + | |
| − | fastx_clipper -h
| + | |
| − | </pre>
| + | |
| − | | + | |
| − | '''Note 1:''' It is good to check the number of reads by running the command:
| + | |
| − | | + | |
| − | <pre>
| + | |
| − | | + | |
| − | wc -l *.trim.fastq
| + | |
| − | | + | |
| − | </pre>
| + | |
| − | | + | |
| − | ==Collapse exact duplicates== | + | |
| − | | + | |
| − | If multiple reads have exactly the same sequence, only one is kept.
| + | |
| − | | + | |
| − | <pre>
| + | |
| − | | + | |
| − | #!/bin/bash
| + | |
| − | #$ -t 1-13 -m a -cwd -N CLIP
| + | |
| − | files=(Fox1_1 Fox1_2 Fox1_3 Fox1_4 Fox2_1 Fox2_2 Fox2_3 Fox2_4 Fox3_1 Fox3_2 Fox3_3 Fox3_4 Fox3_5);
| + | |
| − | f=${files[$SGE_TASK_ID-1]}
| + | |
| − | | + | |
| − | perl /usr/local/CTK/fastq2collapse.pl $f.trim.fastq $f.trim.c.fastq
| + | |
| − | | + | |
| − | </pre>
| + | |
| − | | + | |
| − | *Usage and additional explanation of [[CTK_usage#fastq2collapse.pl|fastq2collapse.pl]].
| + | |
| − | | + | |
| − | '''Note 1:''' It is good to check the number of reads by running the command:
| + | |
| − | | + | |
| − | <pre>
| + | |
| − | wc -l *.trim.c.fastq
| + | |
| − | </pre>
| + | |
| − | | + | |
| − | ==Strip barcode==
| + | |
| − | | + | |
| − | The following script removes the 5' degenerate barcode.
| + | |
| − | | + | |
| − | <pre>
| + | |
| − |
| + | |
| − | #The barcode for standard CLIP is 5 nucleotides long
| + | |
| − | for f in Fox1_1 Fox2_1 Fox3_1
| + | |
| − | do
| + | |
| − | perl /usr/local/CTK/stripBarcode.pl -format fastq -len 5 $f.trim.c.fastq $f.trim.c.tag.fastq
| + | |
| − | done
| + | |
| − | | + | |
| − | #the barcode for BrdU CLIP is 14 nucleotides long
| + | |
| − | for f in Fox1_2 Fox1_3 Fox1_4 Fox2_2 Fox2_3 Fox2_4 Fox3_2 Fox3_3 Fox3_4 Fox3_5
| + | |
| − | do
| + | |
| − | perl /usr/local/CTK/stripBarcode.pl -format fastq -len 14 $f.trim.c.fastq $f.trim.c.tag.fastq
| + | |
| − | done
| + | |
| − | </pre>
| + | |
| − | | + | |
| − | | + | |
| − | '''Note 1:''' We include sample index as part of the random barcode here.
| + | |
| − | | + | |
| − | *Usage and additional explanation of [[CTK_usage#stripBarcode.pl|stripBarcode.pl]].
| + | |
| − | | + | |
| − | | + | |
| − | After this step, one can get the distribution of tag length for diagnostic purposes.
| + | |
| − | | + | |
| − | <pre>
| + | |
| − | | + | |
| − | awk '{if(NR%4==2) {print $0}}' $f.trim.c.tag.fastq | awk '{print length($0)}' | sort -n | uniq -c | awk '{print $2"\t"$1}' > $f.trim.c.tag.seqlen.stat.txt
| + | |
| − | | + | |
| − | </pre>
| + | |
| − | | + | |
| − | [http://zhanglab.c2b2.columbia.edu/data/CTK/preprocess.tar.gz You can download fully preprocessed FASTQ files here ] or alternatively use command:
| + | |
| − | | + | |
| − | <pre>
| + | |
| − | wget http://zhanglab.c2b2.columbia.edu/data/CTK/preprocess.tar.gz
| + | |
| − | tar -zxvf preprocess.tar.gz
| + | |
| − | </pre>
| + | |
| − | | + | |
| − | =Read mapping & parsing=
| + | |
| | | | |
| | We are now using BWA (version 0.7.12) for alignment instead of novoalign for two reasons: | | We are now using BWA (version 0.7.12) for alignment instead of novoalign for two reasons: |
| − | * novoalign is slower than some of the other algorithms that becomes available, in part because the academic version of novoalign does not allow multi threading. | + | * novoalign is slower than some of the other algorithms that become available, in part because the academic version of novoalign does not allow multi threading. |
| | * BWA allows one to specify mismatch rate instead of the the absolute number, which is more appropriate for tags of different sizes (i.e. a smaller number of mismatches allowed for shorter tags after trimming). | | * BWA allows one to specify mismatch rate instead of the the absolute number, which is more appropriate for tags of different sizes (i.e. a smaller number of mismatches allowed for shorter tags after trimming). |
| − |
| |
| − | ==Indexing reference genome ==
| |
| | | | |
| | This step needs to be done only once. | | This step needs to be done only once. |
| Line 538: |
Line 214: |
| | <pre> | | <pre> |
| | cat ch1.fa chr2.fa chr3.fa chr4.fa chr5.fa chr6.fa chr7.fa chr8.fa chr9.fa chr10.fa chr11.fa chr12.fa chr13.fa chr14.fa chr15.fa chr16.fa chr17.fa chr18.fa chr19.fa chr20.fa chr21.fa chr22.fa chrX.fa chrY.fa > mm10.fa | | cat ch1.fa chr2.fa chr3.fa chr4.fa chr5.fa chr6.fa chr7.fa chr8.fa chr9.fa chr10.fa chr11.fa chr12.fa chr13.fa chr14.fa chr15.fa chr16.fa chr17.fa chr18.fa chr19.fa chr20.fa chr21.fa chr22.fa chrX.fa chrY.fa > mm10.fa |
| − | #make sure each chromosome is named by '>chrN' instead of '>N'
| |
| − | #We do not include random chromosomes or chrM.
| |
| | | | |
| − | </pre>
| |
| | | | |
| − | Finally, create a BWA index and move it to a directory you like. In this example, the index is in the /genomes/mm10/bwa/ directory.
| |
| − | <pre>
| |
| − | cd /genomes/mm10/bwa/
| |
| − | bwa index -a bwtsw mm10.fa
| |
| | </pre> | | </pre> |
| | | | |
| − | ==Read mapping==
| + | '''Note 1''' : Make sure each chromosome is named by '>chrN' instead of '>N' |
| − | Now that the reference index has been prepared, you can proceed with mapping/alignment.
| + | |
| | | | |
| − | <pre>
| + | '''Note 2''': In our analysis, we do not include random chromosomes or chrM, which might not be the best for certain projects. |
| | | | |
| − | cd ~/test
| |
| − | mkdir mapping
| |
| − | cd mapping
| |
| | | | |
| − | </pre>
| + | Finally, create a BWA index and move it to a directory you like. In this example, the index is in the /genomes/mm10/bwa/ directory. |
| − | | + | |
| − | Run bwa to align the reads to the reference genome.
| + | |
| | <pre> | | <pre> |
| − | #!/bin/bash
| + | cd /genomes/mm10/bwa/ |
| − | #$ -t 1-13 -m a -cwd -N CLIP
| + | bwa index -a bwtsw mm10.fa |
| − | files=(Fox1_1 Fox1_2 Fox1_3 Fox1_4 Fox2_1 Fox2_2 Fox2_3 Fox2_4 Fox3_1 Fox3_2 Fox3_3 Fox3_4 Fox3_5);
| + | |
| − | f=${files[$SGE_TASK_ID-1]}
| + | |
| − | | + | |
| − | bwa aln -t 4 -n 0.06 -q 20 /genomes/mm10/bwa/mm10.fa ../filtering/$f.trim.c.tag.fastq > $f.sai
| + | |
| − | bwa samse /genomes/mm10/bwa/mm10.fa $f.sai ../filtering/$f.trim.c.tag.fastq > $f.sam | + | |
| − | | + | |
| − | #the two steps can also be combined, using the following command:
| + | |
| − | #bwa aln -t 4 -n 0.06 -q 20 /genomes/mm10/bwa/mm10.fa ../filtering/$f.trim.c.tag.fastq | bwa samse /genomes/mm10/bwa/mm10.fa - ../filtering/$f.trim.c.tag.fastq > $f.sam
| + | |
| − | | + | |
| | </pre> | | </pre> |
| | | | |
| − | The option -n 0.06 specifies slightly more stringent criteria than the default. The number of allowed mismatches (substitutions or indels) depending on read length is as follows:
| |
| − | <pre>
| |
| | | | |
| − | [bwa_aln] 17bp reads: max_diff = 1
| + | =Genome assemblies with annotations included in CTK= |
| − | [bwa_aln] 20bp reads: max_diff = 2
| + | While CTK is not limited to specific species/genome assemblies in general, several steps require gene annotations. Currently the annotation files of the following assemblies have been included as part of CTK: |
| − | [bwa_aln] 45bp reads: max_diff = 3
| + | *hg38 |
| − | [bwa_aln] 73bp reads: max_diff = 4
| + | *hg19 |
| − | [bwa_aln] 104bp reads: max_diff = 5
| + | *mm10 |
| − | [bwa_aln] 137bp reads: max_diff = 6
| + | *dm6 |
| − | [bwa_aln] 172bp reads: max_diff = 7
| + | |
| − | [bwa_aln] 208bp reads: max_diff = 8
| + | |
| − | [bwa_aln] 244bp reads: max_diff = 9
| + | |
| | | | |
| − | </pre>
| + | If you are interested in certain genome assemblies not currently supported by CTK, feel free to let us know by posting in our [https://groups.google.com/forum/#!forum/ctk-user-group Google CTK user group]. |
| | | | |
| − | The -q option is used to trim low quality reads (an average of 20 or below). However, this does not seem to do too much after the trimming steps above.
| |
| | | | |
| − | ==Parsing SAM file==
| |
| | | | |
| − | <pre>
| |
| − |
| |
| − | #!/bin/bash
| |
| − | #$ -t 1-13 -m a -cwd -N CLIP
| |
| − | files=(Fox1_1 Fox1_2 Fox1_3 Fox1_4 Fox2_1 Fox2_2 Fox2_3 Fox2_4 Fox3_1 Fox3_2 Fox3_3 Fox3_4 Fox3_5);
| |
| − | f=${files[$SGE_TASK_ID-1]}
| |
| − |
| |
| − | perl /usr/local/CTK/parseAlignment.pl -v --map-qual 1 --min-len 18 --mutation-file $f.mutation.txt $f.sam $f.tag.bed
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − |
| |
| − | This will keep only unique mappings (with MAPQ >=1) and a minimal mapping size of 18 nt.
| |
| − |
| |
| − | '''Note 1:''' In the tag bed file, the 5′ column records the number of mismatches (substitutions) in each read
| |
| − |
| |
| − | '''Note 2:''' Other aligners might not use a positive MAPQ as an indication of unique mapping.
| |
| − |
| |
| − | Another useful option is --indel-to-end, which specifies the number of nucleotides towards the end from which indels should not be called (default=5 nt).
| |
| − |
| |
| − | '''Note 3:''' The parsing script relies on MD tags, which is an optional field without strict definition in SAM file format specification. Some aligners might have slightly different format how they report mismatches. If other aligners than bwa is used, one should run the following command:
| |
| − |
| |
| − | <pre>
| |
| − |
| |
| − | samtools view -bS $f.sam | samtools sort - $f.sorted
| |
| − | samtools fillmd $f.sorted.bam /genomes/mm10/bwa/mm10.fa > $f.sorted.md.sam
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − | This will ensure the sam file gets parsed properly.
| |
| − |
| |
| − | *Usage and additional explanation of [[CTK_usage#parseAlignment.pl|parseAlignment.pl]].
| |
| − |
| |
| − |
| |
| − | '''Note 4:''' Keep track what proportion of reads can be mapped uniquely.
| |
| − | <pre>
| |
| − |
| |
| − | wc -l *.tag.bed
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − | '''Note 4:''' the format of the mutation file is essentially the same as before.
| |
| − |
| |
| − | =Collapsing PCR duplicates=
| |
| − |
| |
| − | It is critical to collapse PCR duplicates, not only for the exact duplicates collapsed above, but also for those with slight differences due to sequencing errors.
| |
| − |
| |
| − | <pre>
| |
| − | #!/bin/bash
| |
| − | #$ -t 1-13 -m a -cwd -N CLIP
| |
| − | files=(Fox1_1 Fox1_2 Fox1_3 Fox1_4 Fox2_1 Fox2_2 Fox2_3 Fox2_4 Fox3_1 Fox3_2 Fox3_3 Fox3_4 Fox3_5);
| |
| − | f=${files[$SGE_TASK_ID-1]}
| |
| − |
| |
| − | perl /usr/local/CTK/tag2collapse.pl -v -big --random-barcode -EM 30 --seq-error-model alignment -weight --weight-in-name --keep-max-score --keep-tag-name $f.tag.bed $f.tag.uniq.bed
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − | *Usage and additional explanation of [[CTK_usage#tag2collapse.pl|tag2collapse.pl]].
| |
| − |
| |
| − | In the command above, a model-based algorithm is used to identify "sufficiently distinct" barcodes. Details of the algorithm was described in the following paper:
| |
| − |
| |
| − | <pre>
| |
| − | Darnell JC, et al. FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell. 2011; 146:247–261.
| |
| − | </pre>
| |
| − |
| |
| − | Compared to the original algorithm, the current implementation estimates sequencing error from aligned reads, which is then fixed during the iterative EM procedure.
| |
| − |
| |
| − |
| |
| − | '''Note 1:''' Sequencing errors in the degenerate barcodes are estimated from results of read alignment. In addition, the number of substitutions must be provided in the 5th column. Note that the read ID in the 4th column must take the form READ#x#NNNNN, where x is the number of exact duplicates and NNNNN is the bar-code nucleotide sequence appended to read IDs in previous steps. Read IDs that are not in this format will generate an error.
| |
| − |
| |
| − |
| |
| − | Get the mutations in unique tags
| |
| − | <pre>
| |
| − |
| |
| − | #!/bin/bash
| |
| − | #$ -t 1-13 -m a -cwd -N CLIP
| |
| − | files=(Fox1_1 Fox1_2 Fox1_3 Fox1_4 Fox2_1 Fox2_2 Fox2_3 Fox2_4 Fox3_1 Fox3_2 Fox3_3 Fox3_4 Fox3_5);
| |
| − | f=${files[$SGE_TASK_ID-1]}
| |
| − |
| |
| − | perl /usr/local/CTK/selectRow.pl -q 3 -f 3 $f.mutation.txt $f.tag.uniq.bed > $f.tag.uniq.mutation.txt
| |
| − |
| |
| − | #or alternatively, use the following python script
| |
| − | #python /usr/local/CTK/joinWrapper.py $f.mutation.txt $f.tag.uniq.bed 4 4 N $f.tag.uniq.mutation.txt
| |
| − | #the parameters 4 4 indicate the columns in the two input file used to join the two, and N indicates that only paired rows should be printed
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − | *Usage and additional explanation of [[CTK_usage#selectRow.pl|selectRow.pl]].
| |
| − |
| |
| − | Table 2 summarizes the number of unique mutations of different types in each sample.
| |
| − |
| |
| − | {| class="wikitable"
| |
| − | |+Table 2: Summary of mutations in unique tags.
| |
| − | ! Protein
| |
| − | ! Sample
| |
| − | ! CLIP protocol
| |
| − | ! # of unique tags
| |
| − | ! Deletions
| |
| − | ! Insertions
| |
| − | ! Substitutions
| |
| − | |-
| |
| − | | Fox1
| |
| − | | Fox1_1
| |
| − | | Standard
| |
| − | | 357,094
| |
| − | | 30,675
| |
| − | | 4,705
| |
| − | | 112,837
| |
| − | |-
| |
| − | | Fox1
| |
| − | | Fox1_2
| |
| − | | BrdU
| |
| − | | 94,757
| |
| − | | 2,742
| |
| − | | 2,022
| |
| − | | 29,840
| |
| − | |-
| |
| − | | Fox1
| |
| − | | Fox1_3
| |
| − | | BrdU
| |
| − | | 161,487
| |
| − | | 7,348
| |
| − | | 3,138
| |
| − | | 47,372
| |
| − | |-
| |
| − | | Fox1
| |
| − | | Fox1_4
| |
| − | | BrdU
| |
| − | | 1,194,617
| |
| − | | 43,953
| |
| − | | 11,012
| |
| − | | 342,695
| |
| − | |-
| |
| − | | Fox2
| |
| − | | Fox2_1
| |
| − | | Standard
| |
| − | | 295,180
| |
| − | | 26,791
| |
| − | | 6,134
| |
| − | | 103,992
| |
| − | |-
| |
| − | | Fox2
| |
| − | | Fox2_2
| |
| − | | BrdU
| |
| − | | 83,358
| |
| − | | 1,192
| |
| − | | 1,832
| |
| − | | 25,128
| |
| − | |-
| |
| − | | Fox2
| |
| − | | Fox2_3
| |
| − | | BrdU
| |
| − | | 337,811
| |
| − | | 10,911
| |
| − | | 3,442
| |
| − | | 75,912
| |
| − | |-
| |
| − | | Fox2
| |
| − | | Fox2_4
| |
| − | | BrdU
| |
| − | | 165,645
| |
| − | | 2,292
| |
| − | | 5,009
| |
| − | | 67,585
| |
| − | |-
| |
| − | | Fox3
| |
| − | | Fox3_1
| |
| − | | Standard
| |
| − | | 877,256
| |
| − | | 51,536
| |
| − | | 13,896
| |
| − | | 27,1740
| |
| − | |-
| |
| − | | Fox3
| |
| − | | Fox3_2
| |
| − | | BrdU
| |
| − | | 143,522
| |
| − | | 6,058
| |
| − | | 2,059
| |
| − | | 34,729
| |
| − | |-
| |
| − | | Fox3
| |
| − | | Fox3_3
| |
| − | | BrdU
| |
| − | | 399,878
| |
| − | | 17,004
| |
| − | | 3,499
| |
| − | | 73,882
| |
| − | |-
| |
| − | | Fox3
| |
| − | | Fox3_4
| |
| − | | BrdU
| |
| − | | 413,847
| |
| − | | 17,195
| |
| − | | 3,153
| |
| − | | 72,674
| |
| − | |-
| |
| − | | Fox3
| |
| − | | Fox3_5
| |
| − | | BrdU
| |
| − | | 1,076,782
| |
| − | | 38,700
| |
| − | | 10,790
| |
| − | | 374,446
| |
| − | |-
| |
| − | | '''Total'''
| |
| − | |
| |
| − | |
| |
| − | | '''5,601,234'''
| |
| − | | '''256,397'''
| |
| − | | '''70,691'''
| |
| − | | '''1,632,832'''
| |
| − | |}
| |
| − |
| |
| − | [http://zhanglab.c2b2.columbia.edu/data/CTK/unique.tar.gz You can download unique tag and mutation files here.]
| |
| − |
| |
| − | After getting the unique tags of each library, one might concatenate biological replicates, which are distinguished by different colors. As an example:
| |
| − |
| |
| − | <pre>
| |
| − |
| |
| − | perl /usr/local/CTK/bed2rgb.pl -v -col "128,0,0" Fox1_1.tag.uniq.bed Fox1_1.tag.uniq.rgb.bed #as one example
| |
| − |
| |
| − | #repeat the above step for all other uniq.bed files to generate rgb.bed files, but use different rgb colors "x,x,x" (see http://www.rapidtables.com/web/color/RGB_Color.htm or other charts)
| |
| − |
| |
| − | cat *tag.uniq.rgb.bed > Fox.pool.tag.uniq.rgb.bed
| |
| − | cat *tag.uniq.mutation.txt > Fox.pool.tag.uniq.mutation.txt
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − | *Usage and additional explanation of [[CTK_usage#bed2rgb.pl|bed2rgb.pl]].
| |
| − |
| |
| − |
| |
| − | We also concatenate the BrdU CLIP BED files separately for CITS analysis downstream in this pipeline.
| |
| − |
| |
| − | <pre>
| |
| − |
| |
| − | cat Fox1_2.tag.uniq.rgb.bed Fox1_3.tag.uniq.rgb.bed Fox1_4.tag.uniq.rgb.bed Fox2_2.tag.uniq.rgb.bed Fox2_3.tag.uniq.rgb.bed Fox2_4.tag.uniq.rgb.bed Fox3_2.tag.uniq.rgb.bed Fox3_3.tag.uniq.rgb.bed Fox3_4.tag.uniq.rgb.bed Fox3_5.tag.uniq.rgb.bed > Fox_BrdU.pool.tag.uniq.rgb.bed
| |
| − |
| |
| − |
| |
| − | cat Fox1_2.tag.uniq.mutation.txt Fox1_3.tag.uniq.mutation.txt Fox1_4.tag.uniq.mutation.txt Fox2_2.tag.uniq.mutation.txt Fox2_3.tag.uniq.mutation.txt Fox2_4.tag.uniq.mutation.txt Fox3_2.tag.uniq.mutation.txt Fox3_3.tag.uniq.mutation.txt Fox3_4.tag.uniq.mutation.txt Fox3_5.tag.uniq.mutation.txt > Fox_BrdU.pool.tag.uniq.mutation.txt
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − | '''Note 1:''' As a diagnostic step, get the length distribution of unique tags, which should be a more faithful representation of the library:
| |
| − |
| |
| − | <pre>
| |
| − | for f in Fox1_1 Fox1_2 Fox1_3 Fox1_4 Fox2_1 Fox2_2 Fox2_3 Fox2_4 Fox3_1 Fox3_2 Fox3_3 Fox3_4 Fox3_5
| |
| − | do
| |
| − | awk '{print $3-$2}' $f.tag.uniq.bed | sort -n | uniq -c | awk '{print $2"\t"$1}' > $f.uniq.len.dist.txt
| |
| − | done
| |
| − | </pre>
| |
| − |
| |
| − | '''Note 2:''' get genomic distribution of CLIP tags
| |
| − |
| |
| − | <pre>
| |
| − | perl /usr/local/CTK/bed2annotation.pl -dbkey mm10 -ss -big -region -v -summary Fox.pool.tag.uniq.annot.summary.txt Fox.pool.tag.uniq.rgb.bed Fox.pool.tag.uniq.annot.txt
| |
| − | </pre>
| |
| − |
| |
| − | Make sure the current genome (mm10) is specified (mm10 and hg19 are currently supported).
| |
| − |
| |
| − | Check the summary file (<tag.uniq.annot.summary.txt>) for the percentage of tags mapped to CDS, 3'UTR, introns, etc.
| |
| − |
| |
| − | *Usage and additional explanation of [[CTK_usage#bed2annotation.pl|bed2annotation.pl]].
| |
| − |
| |
| − | '''Note 3:''' generate bedgraph for visualization in the genome browser:
| |
| − |
| |
| − | <pre>
| |
| − | perl /usr/local/CTK/tag2profile.pl -v -ss -exact -of bedgraph -n ″Unique Tag Profile″ Fox.pool.tag.uniq.rgb.bed Fox.pool.tag.uniq.bedgraph
| |
| − | </pre>
| |
| − |
| |
| − | *Usage and additional explanation of [[CTK_usage#tag2profile.pl|tag2profile.pl]].
| |
| − |
| |
| − | =Peak calling=
| |
| − |
| |
| − | <pre>
| |
| − | cd ~/test
| |
| − | mkdir cluster
| |
| − | cd cluster
| |
| − | ln -s ../mapping/Fox.pool.tag.uniq.rgb.bed
| |
| − | </pre>
| |
| − |
| |
| − | CTK provides two modes of peak calling. The first mode is peak calling without statistical assessment, which will find all potential peaks. The second mode is to find peaks with peak height more than one expected by chance.
| |
| − |
| |
| − | ==Mode 1: Peak calling without statistical assessment==
| |
| − | <pre>
| |
| − |
| |
| − | perl /usr/local/CTK/tag2peak.pl -big -ss -v --valley-seeking --valley-depth 0.9 Fox.pool.tag.uniq.bed Fox.pool.tag.uniq.peak.bed --out-boundary Fox.pool.tag.uniq.peak.boundary.bed --out-half-PH Fox.pool.tag.uniq.peak.halfPH.bed
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − | This script searches for all potential peaks using a "valley seeking" algorithm. A peak is called if valleys of certain depths are found on both sides so that it is separated from other peaks. The results are similar to those generated by the original clustering algorithm. However, this valley seeking algorithm is advantageous because it is able to separate neighboring peaks without zero-tag gaps.
| |
| − |
| |
| − |
| |
| − | '''Note 1:''' To annotate peaks with overlapping genes and repeat masked sequences and get genomic breakdown:
| |
| − |
| |
| − | <pre>
| |
| − | perl /usr/local/CTK/bed2annotation.pl -dbkey mm10 -ss -big -region -v -summary Fox.pool.tag.uniq.peak.annot.summary.txt Fox.pool.tag.uniq.peak.bed Fox.pool.tag.uniq.peak.annot.txt
| |
| − | </pre>
| |
| − |
| |
| − | The output file Fox.pool.tag.uniq.peak.annot.txt has the detailed annotation for each peak, and Fox.pool.tag.uniq.peak.annot.summary.txt has summary statistics.
| |
| − |
| |
| − | '''Note 1:''' Another useful option --valley-depth specifies the depth of the valley relative to the peak (0-1, 0.5 by default). One also has the option to merge peaks close to each other by specifying the distance between the peaks (e.g., -gap 20).
| |
| − |
| |
| − | '''Note 2:''' Besides the peak boundaries (Fox.pool.tag.uniq.peak.bed), one can also output cluster boundaries (--out-boundary) and half peak boundaries (--out-half-PH ) associated with each peak.
| |
| − |
| |
| − | The default is set to a valley depth of 0.9.
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − | In this mode, the script will not assess the statistical significance of the peak height. If one needs this, an alternative mode is as follows:
| |
| − |
| |
| − | ==Mode 2: Peak calling with statistical assessment==
| |
| − |
| |
| − | <pre>
| |
| − |
| |
| − | perl /usr/local/CTK/tag2peak.pl -big -ss -v --valley-seeking -p 0.05 --valley-depth 0.9 --dbkey mm10 --multi-test Fox.pool.tag.uniq.bed Fox.pool.tag.uniq.peak.sig.bed --out-boundary Fox.pool.tag.uniq.peak.sig.boundary.bed --out-half-PH Fox.pool.tag.uniq.peak.sig.halfPH.bed
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − | Here, the script searches for significant peaks by considering one specific genic region as a unit for permutations to estimate expected peak height (scan statistics is used here, so the permutation is only conceptual).
| |
| − |
| |
| − | It is important to note that in this mode, only clusters overlapping with annotated genes are kept.
| |
| − |
| |
| − | In both cases, the three output files provide the positions of peak (many times it is 1nt, but not always), the peak width at half peak height, and the boundaries of each peak. In practice, it appears that the center of peak or half-peak provides the most robust/precise indicator of the binding sites (although a shift might be present because of CITS, which can be adjusted after one examines the enrichment of the motif).
| |
| − |
| |
| − | <pre>
| |
| − | wc -l Fox.pool.tag.uniq.peak.sig.bed
| |
| − | 29059 Fox.pool.tag.uniq.peak.sig.bed
| |
| − | </pre>
| |
| − |
| |
| − | '''Note 1:''' To search for a known binding motif, one first defines the center of each peak (based on width at half PH).
| |
| − |
| |
| − | <pre>
| |
| − |
| |
| − | awk '{print $1"\t"int(($2+$3)/2)-500"\t"int(($2+$3)/2)+500"\t"$4"\t"$5"\t"$6}' Fox.pool.tag.uniq.peak.sig.halfPH.bed > Fox.pool.tag.uniq.peak.sig.halfPH.center.ext1k.bed
| |
| − | #+/-500 around peak center
| |
| − | </pre>
| |
| − |
| |
| − | This bed file can be used to extract sequences around peak (pay attention to the strand), and search for enrichment of specific motif (e.g., UGCAUG) relative to the peak.
| |
| − |
| |
| − | '''Note 2:'''One might want to count the number of tags overlapping with each cluster/peak for each sample (e.g., to evaluate correlation between replicates).
| |
| − |
| |
| − | <pre>
| |
| − | perl /usr/local/CTK/tag2profile.pl -ss -region Fox.pool.tag.uniq.peak.sig.boundary.bed -of bed -v Fox.tag.uniq.bed Fox.tag.uniq.peak.sig.boundary.count.bed
| |
| − | </pre>
| |
| − |
| |
| − |
| |
| − |
| |
| − | [http://zhanglab.c2b2.columbia.edu/data/CTK/peak_calling.tar.gz Download input and output files for peak calling here.]
| |
| − |
| |
| − | =CIMS analysis=
| |
| − |
| |
| − |
| |
| − |
| |
| − | Here we go back to the individual Fox files, such as Fox1_1.tag.uniq.bed and Fox1_1.tag.uniq.mutation.txt, and symbolically link them here:
| |
| − |
| |
| − | <pre>
| |
| − | cd ~/test
| |
| − | mkdir CIMS
| |
| − | cd CIMS
| |
| − | for f in Fox1_1 Fox1_2 Fox1_3 Fox1_4 Fox2_1 Fox2_2 Fox2_3 Fox2_4 Fox3_1 Fox3_2 Fox3_3 Fox3_4 Fox3_5
| |
| − | do
| |
| − | ln -s ../mapping/$f.tag.uniq.mutation.txt ./
| |
| − | ln -s ../mapping/$f.tag.uniq.bed ./
| |
| − | done
| |
| − | </pre>
| |
| − |
| |
| − | ==Get specific types of mutations==
| |
| − | Get specific types of mutations, such as deletions, substitutions, and insertions around the cross-linked mutation site.
| |
| − |
| |
| − | <pre>
| |
| − |
| |
| − | for f in Fox1_1 Fox1_2 Fox1_3 Fox1_4 Fox2_1 Fox2_2 Fox2_3 Fox2_4 Fox3_1 Fox3_2 Fox3_3 Fox3_4 Fox3_5
| |
| − | do
| |
| − | awk '{if($9==">") {print $0}}' $f.tag.uniq.mutation.txt | cut -f 1-6 > $f.tag.uniq.sub.bed
| |
| − | awk '{if($9=="-") {print $0}}' $f.tag.uniq.mutation.txt | cut -f 1-6 > $f.tag.uniq.del.bed
| |
| − | awk '{if($9=="+") {print $0}}' $f.tag.uniq.mutation.txt | cut -f 1-6 > $f.tag.uniq.ins.bed
| |
| − | done
| |
| − |
| |
| − | cat *tag.uniq.sub.bed > Fox.pool.tag.uniq.sub.bed
| |
| − | cat *tag.uniq.del.bed > Fox.pool.tag.uniq.del.bed
| |
| − | cat *tag.uniq.ins.bed > Fox.pool.tag.uniq.ins.bed
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − | It is always a good practice to look at the number of each type of mutation to see, e.g., compare the relative abundance of deletions to insertions.
| |
| − |
| |
| − | For example, to get the number of deletions of different sizes:
| |
| − | <pre>
| |
| − |
| |
| − | awk '{print $3-$2}' Fox.pool.tag.uniq.del.bed | sort -n | uniq -c
| |
| − |
| |
| − | 180787 1
| |
| − | 74788 2
| |
| − | 822 3
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − | ==Get CIMS==
| |
| − |
| |
| − | Here we use deletions as an example.
| |
| − |
| |
| − | <pre>
| |
| − | perl /usr/local/CTK/CIMS.pl -n 10 -p -v --keep-cache -c cache_del Fox.pool.tag.uniq.bed Fox.pool.tag.uniq.del.bed Fox.pool.tag.uniq.del.CIMS.txt
| |
| − | </pre>
| |
| − |
| |
| − | By default, CIMS.pl will output all sites including those that are not statistically significant. This is recommended because one can play with stringency later.
| |
| − |
| |
| − | *Usage and additional explanation of [[CTK_usage#CIMS.pl|CIMS.pl]].
| |
| − |
| |
| − | <pre>
| |
| − |
| |
| − | awk '{if($9<=0.001) {print $0}}' Fox.pool.tag.uniq.del.CIMS.txt | sort -k 9,9n -k 8,8nr -k 7,7n > Fox.pool.tag.uniq.del.CIMS.s30.txt
| |
| − | cut -f 1-6 Fox.pool.tag.uniq.del.CIMS.s30.txt > Fox.pool.tag.uniq.del.CIMS.s30.bed
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − | This will keep only those with FDR<0.001. Significant sites will also be sorted by FDR, then by the number of tags with mutations, and then by the total number of overlapping tags. One might try different thresholds to get a balance of sensitivity/specificity (e.g. as judged from motif enrichment).
| |
| − |
| |
| − | '''Note 1:''' Another parameter that might be useful to improve signal to noise is m/k (i.g., $8/$7 in awk)
| |
| − |
| |
| − | '''Note 2:''' By default, the command line above will analyze mutations of size 1. Sometimes deletion of multiple nucleotides occurs and those will be ignored here. One can analyze mutations of size 2 by specifying -w 2. Substitutions are always reported as a single nucleotide (even when consecutive nucleotides are substituted), and insertions occur technically in one position and are thus treated as size 1.
| |
| − |
| |
| − | '''Note 3:''' To examine enrichment of motif around CIMS;
| |
| − |
| |
| − | <pre>
| |
| − |
| |
| − | awk '{print $1"\t"$2-10"\t"$3+10"\t"$4"\t"$5"\t"$6}' Fox.pool.tag.uniq.del.CIMS.s30.bed > Fox.pool.tag.uniq.del.CIMS.s30.21nt.bed
| |
| − | #+/-10 around CIMS
| |
| − | </pre>
| |
| − |
| |
| − | This BED file could be used to extract sequences around CIMS (pay attention to the strand), and search for enrichment of specific motif (e.g., UGCAUG for the case of Rbfox1-3) relative to the peak.
| |
| − |
| |
| − | [http://zhanglab.c2b2.columbia.edu/data/CTK/CIMS.tar.gz Download CIMS output files here.]
| |
| − |
| |
| − | One can repeat these steps for the other types of mutations (i.e. substitutions and insertions).
| |
| − |
| |
| − | =CITS analysis=
| |
| − |
| |
| − | Only certain variations of the CLIP protocol allow you to perform CITS analysis (e.g. iCLIP, BrdU CLIP, etc).
| |
| − |
| |
| − | Therefore, link only the BrdU CLIP files (i.e. exclude the Standard CLIP: Fox1_1, Fox2_1, Fox3_1).
| |
| − | <pre>
| |
| − | cd ~/test
| |
| − | mkdir CITS
| |
| − | cd CITS
| |
| − | ln -s ../CIMS/Fox1_2.tag.uniq.del.bed ./ #as one example
| |
| − | #repeat the above step for all other uniq.del.bed files
| |
| − |
| |
| − | ln -s ../mapping/Fox1_2.tag.uniq.bed ./
| |
| − | ./ #as one example
| |
| − |
| |
| − | #repeat the above step for all other uniq.bed files
| |
| − |
| |
| − | cat *tag.uniq.del.bed > Fox_BrdU.pool.tag.uniq.del.bed
| |
| − | cat *tag.uniq.bed > Fox_BrdU.pool.tag.uniq.bed
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − | Since the vast majority of deletions are introduced because of cross linking, tags with deletions are treated as read-through tags and removed.
| |
| − |
| |
| − | <pre>
| |
| − | perl /usr/local/CTK/removeRow.pl -q 3 -f 3 -v Fox_BrdU.pool.tag.uniq.bed Fox_BrdU.pool.tag.uniq.del.bed > Fox_BrdU.pool.tag.uniq.clean.bed
| |
| − | </pre>
| |
| − |
| |
| − | *Usage and additional explanation of [[CTK_usage#removeRow.pl|removeRow.pl]].
| |
| − |
| |
| − |
| |
| − | Get the position before the start site as a potential cross link site that causes truncation.
| |
| − |
| |
| − | <pre>
| |
| − |
| |
| − | perl /usr/local/CTK/bedExt.pl -n up -l "-1" -r "-1" -v Fox_BrdU.pool.tag.uniq.clean.bed Fox_BrdU.pool.tag.uniq.clean.trunc.bed
| |
| − |
| |
| − | </pre>
| |
| − | *Usage and additional explanation of [[CTK_usage#bedExt.pl|bedExt.pl]].
| |
| − |
| |
| − |
| |
| − | Cluster overlapping tags.
| |
| − |
| |
| − | <pre>
| |
| − |
| |
| − | perl /usr/local/CTK/tag2cluster.pl -big -s -maxgap "-1" -of bed -v Fox_BrdU.pool.tag.uniq.bed Fox_BrdU.pool.tag.uniq.cluster.0.bed
| |
| − | awk '{if($5>2) {print $0}}' Fox_BrdU.pool.tag.uniq.cluster.0.bed > Fox_BrdU.pool.tag.uniq.cluster.bed
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − |
| |
| − | *Usage and additional explanation of [[CTK_usage#tag2cluster.pl|tag2cluster.pl]].
| |
| − |
| |
| − |
| |
| − | Now we are ready for the CITS analysis:
| |
| − |
| |
| − | <pre>
| |
| − |
| |
| − | perl /usr/local/CTK/tag2peak.pl -big -ss -v --prefix "CITS" -gap 25 -p 0.001 -gene Fox_BrdU.pool.tag.uniq.cluster.bed Fox_BrdU.pool.tag.uniq.clean.trunc.bed Fox_BrdU.pool.tag.uniq.clean.CITS.s30.bed
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − |
| |
| − | '''Note 1:''' One can now perform motif enrichment analysis as described above in the CIMS section.
| |
| − |
| |
| − | '''Note 2:''' In the command line above, we opt to merge sites very close to each other and keep the most significant ones because CITS analysis tends to have more background when read through is frequent.
| |
| − |
| |
| − | <pre>
| |
| − | wc -l Fox_BrdU.pool.tag.uniq.clean.CITS.s30.bed
| |
| − | 115363 Fox_BrdU.pool.tag.uniq.clean.CITS.s30.bed
| |
| − | </pre>
| |
| | | | |
| − | [http://zhanglab.c2b2.columbia.edu/data/CTK/CITS.tar.gz Download CITS output files here.] | + | <center>[[CTK|CTK home]] | [[Standard/BrdU-CLIP data analysis using CTK|Standard/BrdU-CLIP]] | [[iCLIP data analysis using CTK|iCLIP]] | [[eCLIP data analysis using CTK|eCLIP]] | [[PARCLIP data analysis using CTK|PAR-CLIP]] | [[CTK_usage|CTK usage]] | [[CTK_FAQ|FAQ]]</center> |
Crosslinking induced mutation site (CIMS) and cross linking induced truncation site (CITS) analyses are computational methods for CLIP data analysis to determine the exact protein-RNA crosslink sites and thereby map protein-RNA interactions at single-nucleotide resolution. These methods are based on the observation that UV crosslinked amino-acid-RNA adducts can introduce reverse transcription errors, including mutations and premature in cDNAs at a certain frequency, which are captured by sequencing and subsequent comparison of CLIP tags with a reference genome.
For crosslinking induced trunction analysis (CITS) described below, please refer to:
This software is implemented in perl. It also relies on several standard linux/unix tools such as grep, cat, sort, etc. We have tested the software on Cent OS, although it is expected to work on most unix-like systems, including Mac OS X. In addition, several software packages are required by the pipeline for sequence preprocessing and alignment (the version number in our test is also indicated).
Below are the installation instructions for the perl packages CTK and CZPLIB through Anaconda.
If you would like to install a specific platform version e.g. 'noarch', then use the below command:
For ease of installation, we recommend the above commands to be copied to a script setup_ctk.sh. This will also make it easy to include the required steps (See Note below) for restoring the PATH variable if we want to.
Assuming the conda base environment is already setup and activated, run the setup_ctk.sh script from the terminal.
1. At the beginning of the script setup_ctk.sh, include the following:
2. Just before the line "conda activate $myenv", include the following:
3. Just after the line "conda activate $myenv", include the following:
These steps will write the required activate.d and deactivate.d scripts to reset the $PATH variable
Add the library path to the environment variable, so perl can find it.
Add the dir to your $PATH environment variable if you would like.
Finally, some of the scripts will use a cache directory, which is under the working directory by default. One can specify another folder for cache using environment variable (recommended).
We are now using BWA (version 0.7.12) for alignment instead of novoalign for two reasons:
This step needs to be done only once.
Change the chromosome header and combine the chromosomes into a full genome. Note that we exclude random chromosomes and the mitochondria chromosome in our analysis.
While CTK is not limited to specific species/genome assemblies in general, several steps require gene annotations. Currently the annotation files of the following assemblies have been included as part of CTK:
If you are interested in certain genome assemblies not currently supported by CTK, feel free to let us know by posting in our Google CTK user group.