Difference between revisions of "CTK Documentation"
From Zhang Laboratory
(→Collapsing PCR duplicates: removed rmsk, added gene) |
(→Installation) |
||
| (48 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | <center>[[CTK|CTK home]] | [[Standard/BrdU-CLIP data analysis using CTK|Standard/BrdU-CLIP]] | [[iCLIP data analysis using CTK|iCLIP]] | [[eCLIP data analysis using CTK|eCLIP]] | [[PARCLIP data analysis using CTK|PAR-CLIP]] | [[CTK_usage|CTK usage]] | [[CTK_FAQ|FAQ]]</center> | |
| − | + | __TOC__ | |
| − | + | =Introduction= | |
| − | + | [[File:CTK_Pipeline_Overview.png|500px|center]] | |
| − | + | ||
| − | |||
| − | |||
| + | Crosslinking and immunoprecipitation followed by highthroughput sequencing (HITS-CLIP or CLIP-Seq) has now been widely used to map protein-RNA interactions on a genome-wide scale. The CLIP Tool Kit (CTK) is a software package that provides a set of tools for analysis of CLIP data starting from the raw reads generated by the sequencer. It includes pipelines to filter and map reads, collapse PCR duplicates to obtain unique CLIP tags, define CLIP tag clusters and call peaks, and define the exact protein-RNA crosslink sites by CIMS and CITS analysis. This software package is an expanded version of our previous CIMS package. | ||
| − | + | Crosslinking induced mutation site (CIMS) and cross linking induced truncation site (CITS) analyses are computational methods for CLIP data analysis to determine the exact protein-RNA crosslink sites and thereby map protein-RNA interactions at single-nucleotide resolution. These methods are based on the observation that UV crosslinked amino-acid-RNA adducts can introduce reverse transcription errors, including mutations and premature in cDNAs at a certain frequency, which are captured by sequencing and subsequent comparison of CLIP tags with a reference genome. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | If you use the software, please cite: | ||
<pre> | <pre> | ||
| − | + | Shah,A., Qian,Y., Weyn-Vanhentenryck,S.M., Zhang,C. 2017. CLIP Tool Kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics. 33:566-567. | |
| − | + | ||
</pre> | </pre> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | More details of the biochemical and computational aspects of CLIP can be found in the following references: | |
| − | + | ||
<pre> | <pre> | ||
| − | + | Zhang, C. †, Darnell, R.B. † 2011. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat. Biotech. 29:607-614. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | Moore, J.*, Zhang, C.*, Grantman E.C., Mele, A., Darnell, J.C., Darnell, R.B. 2014. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protocols. 9(2):263-93. doi:10.1038/nprot.2014.012. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</pre> | </pre> | ||
| − | + | For crosslinking induced trunction analysis (CITS) described below, please refer to: | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<pre> | <pre> | ||
| − | + | Weyn-Vanhentenryck,S.,M.*, Mele,A.*, Yan,Q.*, Sun,S., Farny,N., Zhang,Z., Xue,C., Herre,M., Silver,P.A., Zhang,M.Q., Krainer,A.R., Darnell,R.B. †, Zhang,C. † 2014. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 6:1139-1152. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</pre> | </pre> | ||
| − | + | =User group= | |
| − | + | For questions/answers, please visit our user group: https://groups.google.com/forum/#!forum/ctk-user-group | |
| − | + | =Versions= | |
| + | *v1.1.3 (12/2018) current | ||
| + | **minor bug fix | ||
| + | *v1.1.2 (08/02/2018) | ||
| + | **update in hg38 annotation files | ||
| + | *v1.1.1 (07/20/2018) | ||
| + | ** bug fix related to the use of dm6 annotation files | ||
| + | *v1.1.0 ( 07/14/2018 ) | ||
| + | ** included support for dm6. | ||
| + | *v1.0.9 ( 06/12/2018 ) | ||
| + | ** minor bug fix | ||
| + | *v1.0.8 ( 05/24/2018 ) | ||
| + | **improved selection of unique CLIP tags. | ||
| + | **improved support for CIMS anlaysis of particular types of substitutions (e.g., T-C for PAR-CLIP). | ||
| + | **a wrapper CITS.pl is included to simplify CITS anlaysis. | ||
| + | **included additional annotations files. | ||
| + | **included support for hg38. | ||
| + | **minor bug fixes | ||
| + | **improved/expanded documentation and tutorials | ||
| + | **Various improvement and bug fixes to improve efficiency and robustness | ||
| + | *v1.0.7 ( 01-16-2017 ) | ||
| + | **fix mac-specific crash | ||
| + | *v1.0.6 ( 01-04-2017 ) | ||
| + | **minor bug fix | ||
| + | *v1.0.5 ( 11-10-2016 ) | ||
| + | **fixed path to annotation files | ||
| + | **have default path to gene bed file for tag2peak.pl | ||
| + | *v1.0.4 ( 10-05-2016 ) | ||
| + | **minor fixes | ||
| + | *v1.0.3 ( 08-08-2016 ) | ||
| + | **improvement in software packaging and usage | ||
| + | *v1.0.0 ( 10-12-2015 ) | ||
| + | **The initial beta release | ||
| − | + | =Download= | |
| − | + | *czplib (perl): a perl library with various functions for genomic/bioinformatic analysis. [https://github.com/chaolinzhanglab/czplib Download from github] | |
| + | *CTK (perl): the core algorithm. [https://github.com/chaolinzhanglab/ctk Download from github] | ||
| − | + | =Prerequisites= | |
| − | + | This software is implemented in perl. It also relies on several standard linux/unix tools such as grep, cat, sort, etc. We have tested the software on Cent OS, although it is expected to work on most unix-like systems, including Mac OS X. In addition, several software packages are required by the pipeline for sequence preprocessing and alignment (the version number in our test is also indicated). | |
| − | + | *FASTX Tool-Kit Version 0.0.13: http://hannonlab.cshl.edu/fastx_toolkit/download.html | |
| + | *cutadapt Version 1.14: https://pypi.python.org/pypi/cutadapt/ (an alternative to FASTX Tool-Kit) | ||
| + | *Burrows Wheeler Aligner (BWA) Version 0.7.12: http://bio-bwa.sourceforge.net/ | ||
| + | *Samtools Version 1.3.1: http://samtools.sourceforge.net | ||
| + | *Perl Version 5.14.3 was used for testing, but we expect that newer versions of Perl will also be compatible: https://www.perl.org/get.html | ||
| + | *Perl library Math::CDF Version 0.1: http://search.cpan.org/~callahan/Math-CDF-0.1/CDF.pm | ||
| − | + | =Installation= | |
| − | + | ==Through anaconda== | |
| − | + | Below are the installation instructions for the perl packages CTK and CZPLIB through Anaconda. | |
| − | + | * Setup the working environment 'ctk' and install all the packages by running the commands below: | |
| − | + | ||
| − | ' | + | |
| − | + | ||
| − | + | ||
<pre> | <pre> | ||
| + | myenv='ctk' | ||
| + | conda create --yes --name $myenv | ||
| − | + | conda activate $myenv | |
| − | + | conda config --env --append channels conda-forge | |
| − | + | conda config --env --append channels bioconda | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | conda install --yes -c chaolinzhanglab ctk | ||
</pre> | </pre> | ||
| − | + | If you would like to install a specific platform version e.g. 'noarch', then use the below command: | |
| − | + | ||
| − | If | + | |
| − | + | ||
<pre> | <pre> | ||
| − | + | conda install --yes -c chaolinzhanglab/noarch ctk | |
| − | + | ||
| − | + | ||
</pre> | </pre> | ||
| − | + | For ease of installation, we recommend the above commands to be copied to a script setup_ctk.sh. This will also make it easy to include the required steps (See '''Note''' below) for restoring the PATH variable if we want to. | |
| − | + | ||
| − | + | ||
| + | Assuming the conda base environment is already setup and activated, run the setup_ctk.sh script from the terminal. | ||
<pre> | <pre> | ||
| − | + | (base)...$source setup_ctk.sh | |
| − | + | ||
| − | + | ||
</pre> | </pre> | ||
| − | |||
| − | + | Now we can proceed to our working directory for performing the CTK analysis. | |
| − | |||
| − | + | '''Acknowledgment Note:''' | |
| + | There is a R wrapper for CTK, called CLIPflexR which can call some other external libraries within R. | ||
| − | + | For details please visit the CLIPflexR webpage: | |
| − | + | https://kathrynrozengagnon.github.io/CLIPflexR/index.html | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | '''Note:''' | |
| − | + | If we need to reset the PATH variable when we do 'conda deactivate' to go to the base environment from the working environment 'ctk', we suggest to include the following steps in the setup_ctk.sh. | |
| − | + | This is a crude way but serves the purpose. | |
| − | + | 1. At the beginning of the script setup_ctk.sh, include the following: | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<pre> | <pre> | ||
| − | + | export CONDA_PATH_RESET=/usr/local/bin/:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:${CONDA_PREFIX}/bin:${CONDA_PREFIX}/condabin | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</pre> | </pre> | ||
| − | + | 2. Just before the line "conda activate $myenv", include the following: | |
| − | + | ||
| − | + | ||
| − | + | ||
<pre> | <pre> | ||
| + | echo "CONDA_PATH_RESET=$CONDA_PATH_RESET" > "${CONDA_PREFIX}/envs/${myenv}/etc/conda/activate.d/${myenv}_env_activate.sh" | ||
| + | echo "export CONDA_PATH_RESET" >> "${CONDA_PREFIX}/envs/${myenv}/etc/conda/activate.d/${myenv}_env_activate.sh" | ||
| − | + | chmod +x "${CONDA_PREFIX}/envs/${myenv}/etc/conda/activate.d/${myenv}_env_activate.sh" | |
| − | + | ||
</pre> | </pre> | ||
| − | + | 3. Just after the line "conda activate $myenv", include the following: | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<pre> | <pre> | ||
| + | echo "PATH=\$CONDA_PATH_RESET" > "${CONDA_PREFIX}/etc/conda/deactivate.d/${myenv}_env_deactivate.sh" | ||
| + | echo "export PATH" >> "${CONDA_PREFIX}/etc/conda/deactivate.d/${myenv}_env_deactivate.sh" | ||
| − | + | chmod +x "${CONDA_PREFIX}/etc/conda/deactivate.d/${myenv}_env_deactivate.sh" | |
| − | + | ||
| − | + | ||
</pre> | </pre> | ||
| − | + | These steps will write the required activate.d and deactivate.d scripts to reset the $PATH variable | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | ==Manual Installation== | ||
| + | * Download and install software packages described in prerequisites. | ||
| + | * Download the czplib perl library files (refer back to '''Download''' section above) | ||
| + | * Decompress and move to whatever directory you like (as an example, we use /usr/local/lib/) | ||
| + | * Replace "x.tgz" below with the version of the package you downloaded | ||
<pre> | <pre> | ||
| − | + | $unzip czplib-1.0.x.zip | |
| − | + | $mv czplib-1.0.x /usr/local/lib/czplib | |
| − | + | ||
</pre> | </pre> | ||
| − | + | Add the library path to the environment variable, so perl can find it. | |
| − | + | ||
<pre> | <pre> | ||
| − | + | export PERL5LIB=/usr/local/lib/czplib | |
| − | + | ||
| − | + | ||
</pre> | </pre> | ||
| − | + | * Download CTK code and likewise decompress and move to whatever directory you like (as an example, we use /usr/local/) | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<pre> | <pre> | ||
| − | + | $unzip ctk-1.0.x.zip | |
| − | + | $mv ctk-1.0.x /usr/local/CTK | |
| − | .. | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</pre> | </pre> | ||
| − | + | Add the dir to your $PATH environment variable if you would like. | |
| − | + | Finally, some of the scripts will use a cache directory, which is under the working directory by default. One can specify another folder for cache using environment variable (recommended). | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | |||
<pre> | <pre> | ||
| − | + | #e.g., add the following lines in .bash_profile | |
| − | + | CACHEHOME=$HOME/cache | |
| + | export CACHEHOME | ||
</pre> | </pre> | ||
| − | + | =Indexing reference genome = | |
| − | + | We are now using BWA (version 0.7.12) for alignment instead of novoalign for two reasons: | |
| + | * novoalign is slower than some of the other algorithms that become available, in part because the academic version of novoalign does not allow multi threading. | ||
| + | * BWA allows one to specify mismatch rate instead of the the absolute number, which is more appropriate for tags of different sizes (i.e. a smaller number of mismatches allowed for shorter tags after trimming). | ||
| − | + | This step needs to be done only once. | |
| − | + | After you have installed BWA, prepare a reference genome: | |
| − | + | ||
| − | + | ||
| + | For example, build a reference mm10 genome. Download the reference genome here: http://ccb.jhu.edu/software/tophat/igenomes.shtml. In this case, make sure you are downloading the "''Mus musculus'' UCSC MM10" reference. | ||
<pre> | <pre> | ||
| − | + | wget ftp://igenome:G3nom3s4u@ussd-ftp.illumina.com/Mus_musculus/UCSC/mm10/Mus_musculus_UCSC_mm10.tar.gz | |
| − | + | tar -xvf Mus_musculus_UCSC_mm10.tar.gz | |
| − | + | cd /Mus_musculus_UCSC_mm10/Mus_musculus/UCSC/mm10/Sequence/Chromosomes | |
</pre> | </pre> | ||
| − | + | Change the chromosome header and combine the chromosomes into a full genome. Note that we exclude random chromosomes and the mitochondria chromosome in our analysis. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<pre> | <pre> | ||
| + | cat ch1.fa chr2.fa chr3.fa chr4.fa chr5.fa chr6.fa chr7.fa chr8.fa chr9.fa chr10.fa chr11.fa chr12.fa chr13.fa chr14.fa chr15.fa chr16.fa chr17.fa chr18.fa chr19.fa chr20.fa chr21.fa chr22.fa chrX.fa chrY.fa > mm10.fa | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</pre> | </pre> | ||
| − | '''Note | + | '''Note 1''' : Make sure each chromosome is named by '>chrN' instead of '>N' |
| − | + | '''Note 2''': In our analysis, we do not include random chromosomes or chrM, which might not be the best for certain projects. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | Finally, create a BWA index and move it to a directory you like. In this example, the index is in the /genomes/mm10/bwa/ directory. | ||
<pre> | <pre> | ||
| − | + | cd /genomes/mm10/bwa/ | |
| − | cd | + | bwa index -a bwtsw mm10.fa |
| − | + | ||
| − | + | ||
</pre> | </pre> | ||
| − | |||
| − | |||
| − | + | =Genome assemblies with annotations included in CTK= | |
| + | While CTK is not limited to specific species/genome assemblies in general, several steps require gene annotations. Currently the annotation files of the following assemblies have been included as part of CTK: | ||
| + | *hg38 | ||
| + | *hg19 | ||
| + | *mm10 | ||
| + | *dm6 | ||
| − | + | If you are interested in certain genome assemblies not currently supported by CTK, feel free to let us know by posting in our [https://groups.google.com/forum/#!forum/ctk-user-group Google CTK user group]. | |
| − | + | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | <center>[[CTK|CTK home]] | [[Standard/BrdU-CLIP data analysis using CTK|Standard/BrdU-CLIP]] | [[iCLIP data analysis using CTK|iCLIP]] | [[eCLIP data analysis using CTK|eCLIP]] | [[PARCLIP data analysis using CTK|PAR-CLIP]] | [[CTK_usage|CTK usage]] | [[CTK_FAQ|FAQ]]</center> | |
Latest revision as of 13:04, 24 August 2022
Contents
Introduction
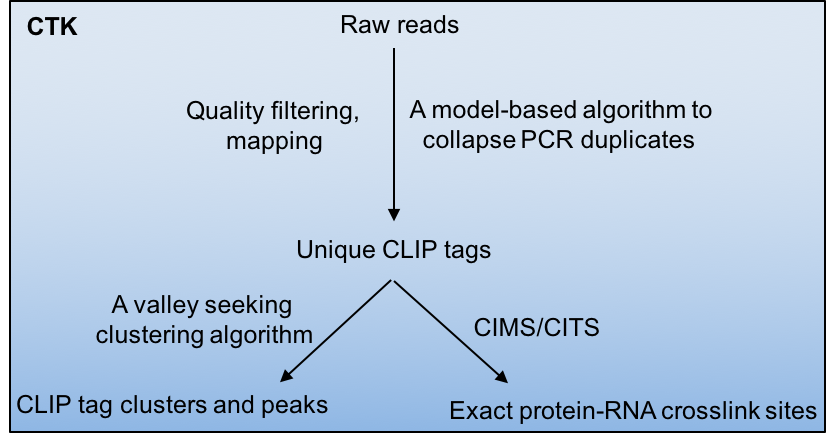
Crosslinking and immunoprecipitation followed by highthroughput sequencing (HITS-CLIP or CLIP-Seq) has now been widely used to map protein-RNA interactions on a genome-wide scale. The CLIP Tool Kit (CTK) is a software package that provides a set of tools for analysis of CLIP data starting from the raw reads generated by the sequencer. It includes pipelines to filter and map reads, collapse PCR duplicates to obtain unique CLIP tags, define CLIP tag clusters and call peaks, and define the exact protein-RNA crosslink sites by CIMS and CITS analysis. This software package is an expanded version of our previous CIMS package.
Crosslinking induced mutation site (CIMS) and cross linking induced truncation site (CITS) analyses are computational methods for CLIP data analysis to determine the exact protein-RNA crosslink sites and thereby map protein-RNA interactions at single-nucleotide resolution. These methods are based on the observation that UV crosslinked amino-acid-RNA adducts can introduce reverse transcription errors, including mutations and premature in cDNAs at a certain frequency, which are captured by sequencing and subsequent comparison of CLIP tags with a reference genome.
If you use the software, please cite:
Shah,A., Qian,Y., Weyn-Vanhentenryck,S.M., Zhang,C. 2017. CLIP Tool Kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics. 33:566-567.
More details of the biochemical and computational aspects of CLIP can be found in the following references:
Zhang, C. †, Darnell, R.B. † 2011. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat. Biotech. 29:607-614. Moore, J.*, Zhang, C.*, Grantman E.C., Mele, A., Darnell, J.C., Darnell, R.B. 2014. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protocols. 9(2):263-93. doi:10.1038/nprot.2014.012.
For crosslinking induced trunction analysis (CITS) described below, please refer to:
Weyn-Vanhentenryck,S.,M.*, Mele,A.*, Yan,Q.*, Sun,S., Farny,N., Zhang,Z., Xue,C., Herre,M., Silver,P.A., Zhang,M.Q., Krainer,A.R., Darnell,R.B. †, Zhang,C. † 2014. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 6:1139-1152.
User group
For questions/answers, please visit our user group: https://groups.google.com/forum/#!forum/ctk-user-group
Versions
- v1.1.3 (12/2018) current
- minor bug fix
- v1.1.2 (08/02/2018)
- update in hg38 annotation files
- v1.1.1 (07/20/2018)
- bug fix related to the use of dm6 annotation files
- v1.1.0 ( 07/14/2018 )
- included support for dm6.
- v1.0.9 ( 06/12/2018 )
- minor bug fix
- v1.0.8 ( 05/24/2018 )
- improved selection of unique CLIP tags.
- improved support for CIMS anlaysis of particular types of substitutions (e.g., T-C for PAR-CLIP).
- a wrapper CITS.pl is included to simplify CITS anlaysis.
- included additional annotations files.
- included support for hg38.
- minor bug fixes
- improved/expanded documentation and tutorials
- Various improvement and bug fixes to improve efficiency and robustness
- v1.0.7 ( 01-16-2017 )
- fix mac-specific crash
- v1.0.6 ( 01-04-2017 )
- minor bug fix
- v1.0.5 ( 11-10-2016 )
- fixed path to annotation files
- have default path to gene bed file for tag2peak.pl
- v1.0.4 ( 10-05-2016 )
- minor fixes
- v1.0.3 ( 08-08-2016 )
- improvement in software packaging and usage
- v1.0.0 ( 10-12-2015 )
- The initial beta release
Download
- czplib (perl): a perl library with various functions for genomic/bioinformatic analysis. Download from github
- CTK (perl): the core algorithm. Download from github
Prerequisites
This software is implemented in perl. It also relies on several standard linux/unix tools such as grep, cat, sort, etc. We have tested the software on Cent OS, although it is expected to work on most unix-like systems, including Mac OS X. In addition, several software packages are required by the pipeline for sequence preprocessing and alignment (the version number in our test is also indicated).
- FASTX Tool-Kit Version 0.0.13: http://hannonlab.cshl.edu/fastx_toolkit/download.html
- cutadapt Version 1.14: https://pypi.python.org/pypi/cutadapt/ (an alternative to FASTX Tool-Kit)
- Burrows Wheeler Aligner (BWA) Version 0.7.12: http://bio-bwa.sourceforge.net/
- Samtools Version 1.3.1: http://samtools.sourceforge.net
- Perl Version 5.14.3 was used for testing, but we expect that newer versions of Perl will also be compatible: https://www.perl.org/get.html
- Perl library Math::CDF Version 0.1: http://search.cpan.org/~callahan/Math-CDF-0.1/CDF.pm
Installation
Through anaconda
Below are the installation instructions for the perl packages CTK and CZPLIB through Anaconda.
- Setup the working environment 'ctk' and install all the packages by running the commands below:
myenv='ctk' conda create --yes --name $myenv conda activate $myenv conda config --env --append channels conda-forge conda config --env --append channels bioconda conda install --yes -c chaolinzhanglab ctk
If you would like to install a specific platform version e.g. 'noarch', then use the below command:
conda install --yes -c chaolinzhanglab/noarch ctk
For ease of installation, we recommend the above commands to be copied to a script setup_ctk.sh. This will also make it easy to include the required steps (See Note below) for restoring the PATH variable if we want to.
Assuming the conda base environment is already setup and activated, run the setup_ctk.sh script from the terminal.
(base)...$source setup_ctk.sh
Now we can proceed to our working directory for performing the CTK analysis.
Acknowledgment Note:
There is a R wrapper for CTK, called CLIPflexR which can call some other external libraries within R.
For details please visit the CLIPflexR webpage:
https://kathrynrozengagnon.github.io/CLIPflexR/index.html
Note: If we need to reset the PATH variable when we do 'conda deactivate' to go to the base environment from the working environment 'ctk', we suggest to include the following steps in the setup_ctk.sh. This is a crude way but serves the purpose.
1. At the beginning of the script setup_ctk.sh, include the following:
export CONDA_PATH_RESET=/usr/local/bin/:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:${CONDA_PREFIX}/bin:${CONDA_PREFIX}/condabin
2. Just before the line "conda activate $myenv", include the following:
echo "CONDA_PATH_RESET=$CONDA_PATH_RESET" > "${CONDA_PREFIX}/envs/${myenv}/etc/conda/activate.d/${myenv}_env_activate.sh"
echo "export CONDA_PATH_RESET" >> "${CONDA_PREFIX}/envs/${myenv}/etc/conda/activate.d/${myenv}_env_activate.sh"
chmod +x "${CONDA_PREFIX}/envs/${myenv}/etc/conda/activate.d/${myenv}_env_activate.sh"
3. Just after the line "conda activate $myenv", include the following:
echo "PATH=\$CONDA_PATH_RESET" > "${CONDA_PREFIX}/etc/conda/deactivate.d/${myenv}_env_deactivate.sh"
echo "export PATH" >> "${CONDA_PREFIX}/etc/conda/deactivate.d/${myenv}_env_deactivate.sh"
chmod +x "${CONDA_PREFIX}/etc/conda/deactivate.d/${myenv}_env_deactivate.sh"
These steps will write the required activate.d and deactivate.d scripts to reset the $PATH variable
Manual Installation
- Download and install software packages described in prerequisites.
- Download the czplib perl library files (refer back to Download section above)
- Decompress and move to whatever directory you like (as an example, we use /usr/local/lib/)
- Replace "x.tgz" below with the version of the package you downloaded
$unzip czplib-1.0.x.zip $mv czplib-1.0.x /usr/local/lib/czplib
Add the library path to the environment variable, so perl can find it.
export PERL5LIB=/usr/local/lib/czplib
- Download CTK code and likewise decompress and move to whatever directory you like (as an example, we use /usr/local/)
$unzip ctk-1.0.x.zip $mv ctk-1.0.x /usr/local/CTK
Add the dir to your $PATH environment variable if you would like.
Finally, some of the scripts will use a cache directory, which is under the working directory by default. One can specify another folder for cache using environment variable (recommended).
#e.g., add the following lines in .bash_profile CACHEHOME=$HOME/cache export CACHEHOME
Indexing reference genome
We are now using BWA (version 0.7.12) for alignment instead of novoalign for two reasons:
- novoalign is slower than some of the other algorithms that become available, in part because the academic version of novoalign does not allow multi threading.
- BWA allows one to specify mismatch rate instead of the the absolute number, which is more appropriate for tags of different sizes (i.e. a smaller number of mismatches allowed for shorter tags after trimming).
This step needs to be done only once.
After you have installed BWA, prepare a reference genome:
For example, build a reference mm10 genome. Download the reference genome here: http://ccb.jhu.edu/software/tophat/igenomes.shtml. In this case, make sure you are downloading the "Mus musculus UCSC MM10" reference.
wget ftp://igenome:G3nom3s4u@ussd-ftp.illumina.com/Mus_musculus/UCSC/mm10/Mus_musculus_UCSC_mm10.tar.gz tar -xvf Mus_musculus_UCSC_mm10.tar.gz cd /Mus_musculus_UCSC_mm10/Mus_musculus/UCSC/mm10/Sequence/Chromosomes
Change the chromosome header and combine the chromosomes into a full genome. Note that we exclude random chromosomes and the mitochondria chromosome in our analysis.
cat ch1.fa chr2.fa chr3.fa chr4.fa chr5.fa chr6.fa chr7.fa chr8.fa chr9.fa chr10.fa chr11.fa chr12.fa chr13.fa chr14.fa chr15.fa chr16.fa chr17.fa chr18.fa chr19.fa chr20.fa chr21.fa chr22.fa chrX.fa chrY.fa > mm10.fa
Note 1 : Make sure each chromosome is named by '>chrN' instead of '>N'
Note 2: In our analysis, we do not include random chromosomes or chrM, which might not be the best for certain projects.
Finally, create a BWA index and move it to a directory you like. In this example, the index is in the /genomes/mm10/bwa/ directory.
cd /genomes/mm10/bwa/ bwa index -a bwtsw mm10.fa
Genome assemblies with annotations included in CTK
While CTK is not limited to specific species/genome assemblies in general, several steps require gene annotations. Currently the annotation files of the following assemblies have been included as part of CTK:
- hg38
- hg19
- mm10
- dm6
If you are interested in certain genome assemblies not currently supported by CTK, feel free to let us know by posting in our Google CTK user group.