Difference between revisions of "Main Page"
From Zhang Laboratory
| Line 177: | Line 177: | ||
* 06/21/2024. Preprint release - Dan Moakley (recent graduate)'s PhD work to infer neuron type-specific splicing-regulatory networks from single-cell RNA-seq transcriptome: http://doi.org/10.1101/2024.06.13.597128. | * 06/21/2024. Preprint release - Dan Moakley (recent graduate)'s PhD work to infer neuron type-specific splicing-regulatory networks from single-cell RNA-seq transcriptome: http://doi.org/10.1101/2024.06.13.597128. | ||
* 06/21/2024. Our MAPT/tau spllcing evolutoin paper is officially out in Cell Genomics. http://www.cell.com/cell-genomics/fulltext/S2666-979X(24)00129-0 | * 06/21/2024. Our MAPT/tau spllcing evolutoin paper is officially out in Cell Genomics. http://www.cell.com/cell-genomics/fulltext/S2666-979X(24)00129-0 | ||
| − | |||
Revision as of 18:32, 25 September 2024
Introduction of the Zhang Laboratory
|
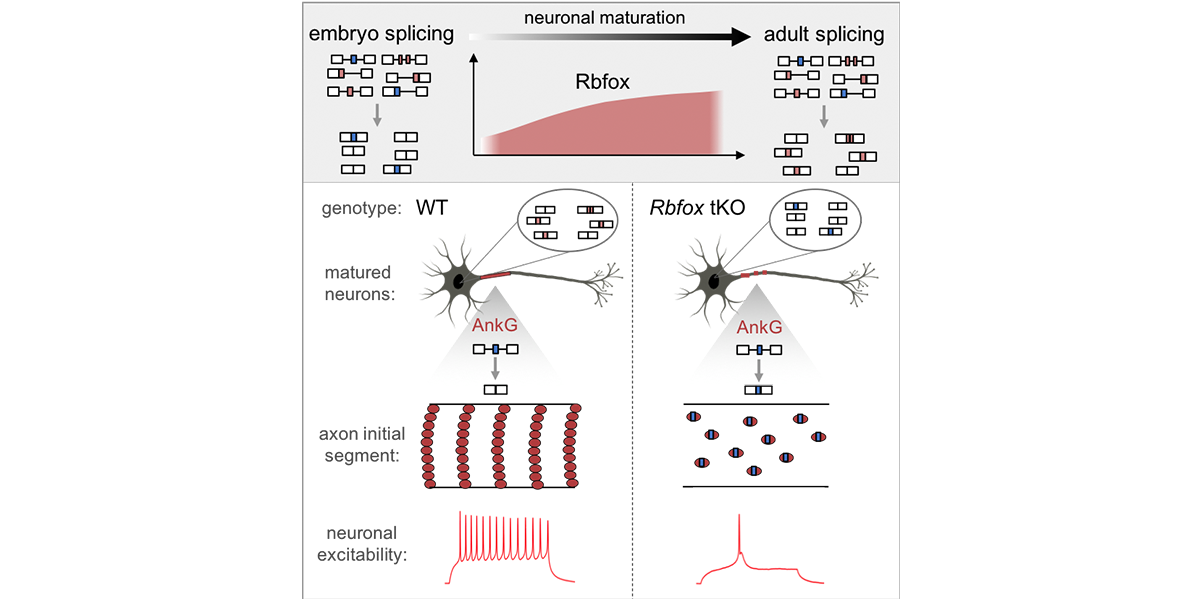
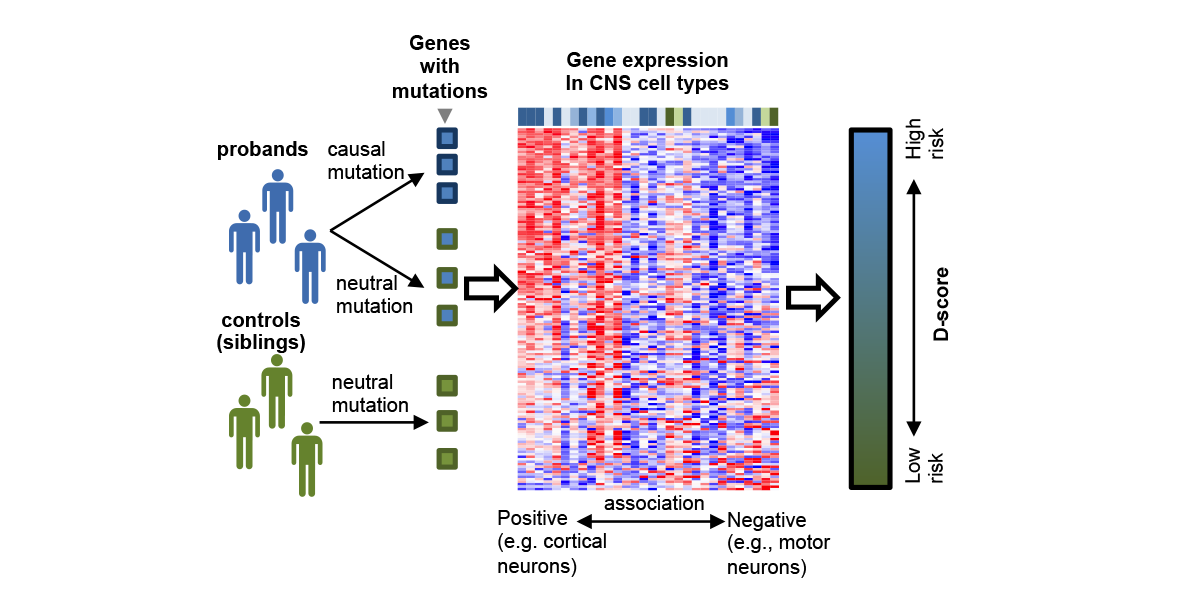
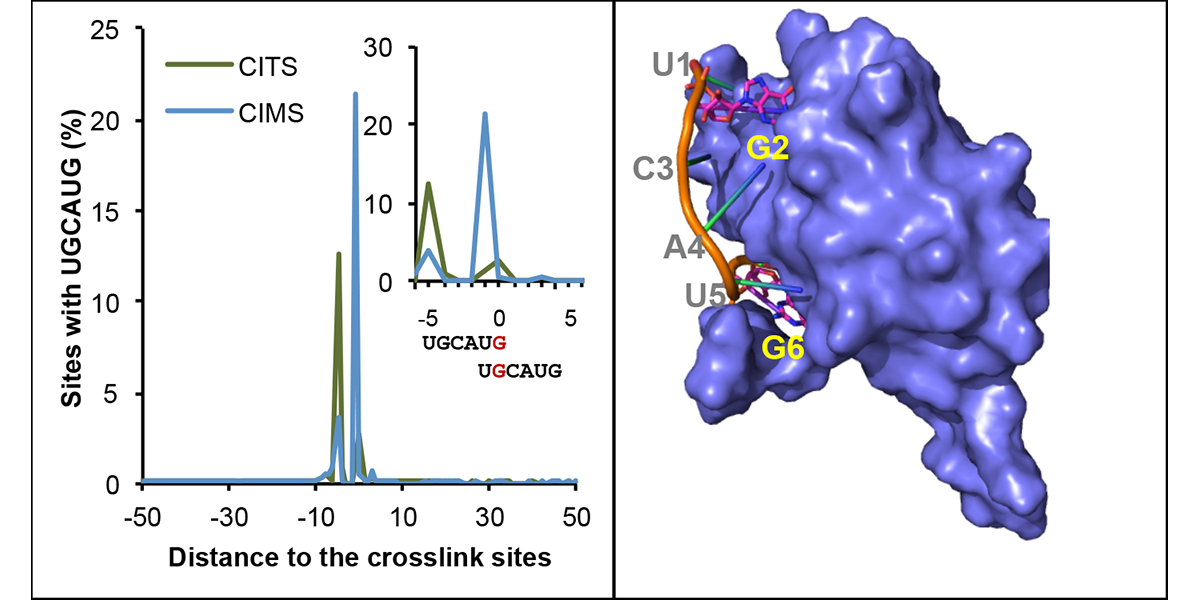
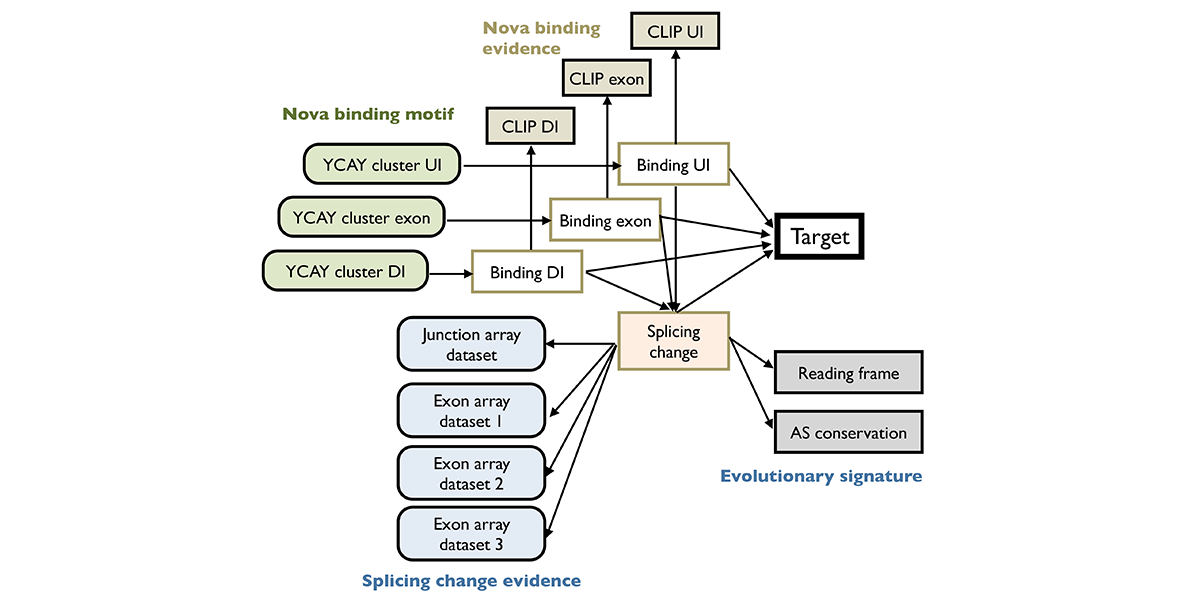
We are part of the Department of Systems Biology, the Department of Biochemistry and Molecular Biophysics, and the Motor Neuron Center at Columbia University Medical Center. We are fascinated by the complexity of the mammalian nervous system and its underlying molecular mechanisms. While mammals have a similar number of genes compared to phenotypically simpler organisms (such as worms), one apparent feature of mammalian genes is their more complicated gene structures, providing an opportunity for sophisticated regulation at the RNA level. The focus of the Zhang lab is to dissect RNA regulatory networks in the nervous system as a way to understand the mammalian complexity manifested in evolutionary-developmental (evo-devo) processes and in several neuronal disorders. The lab has a mixed dry and wet setup, and takes a multidisciplinary approach that tightly integrates high-throughput biochemistry, genomics and computational approaches applied to mouse models and in vitro neuronal differentiation systems from pluripotent stem cells. On the mechanistic side, the Zhang lab focuses on fundamental understanding of the targeting specificity of RNA-binding proteins (RBPs), how they regulate alternative splicing in various cellular contexts, especially in the nervous system, and how such regulation can be disrupted by mutations and genetic variations. On the functional side, the lab aims to uncover the roles of RBPs in determining the neuronal cell fate, morphological and functional properties during neural differentiation and maturation. The lab has also been working on translating fundamental knowledge on RNA regulation to precision genetic medicine, with a particular focus on multiple devastating monogenic diseases affecting the central nervous system.
 2024 Lab Photo - Celebrating Summer Undergrads Amanda Lopez-Ramirez and Chamari Abercrombie, first-row middle and right. (Chamari will stay as a post-bac for the next two years) |
Lab News
|
Chaolin Zhang will co-organize the 2024 Cold Spring Harbor Asia meeting "Computational Biology of the Genome", held in Suzhou China on Oct 21-25. |
>>> all lab news |
Where We Are
|
|
|
|
Columbia University Irving Medical Center (image credit: internet) |
Washington Height, NYC (image credit: @KellyrKopp/twitter) |