What is mCrossBase?
mCrossBase is a database of RNA-binding protein (RBP) binding motifs and crosslink sites defined jointly from CLIP data using a novel algorithm mCross we developed.
Most RBPs recognize very short and degenerate sequence motifs, so defining the precise binidng specificity is challenging, even when CLIP data have become increasingly available. We previously developed a computational approach to map the precise protein-RNA crosslink sites from CLIP data (Zhang & Darnell, Nature Biotechnology, 2011; Weyn-Vanhentenryck et al. Cell Reports, 2014). We also observed that crosslinking frequently occurs at specific positions in RBP binding motifs. To improve the accuracy of de novo RBP motif discovery, we developed a new computational method named mCross that leverages the crosslink sites mapped at the single-nucleotide resolution to precisely register the motif sites, which dramatically reduces the search space (see the workflow below).
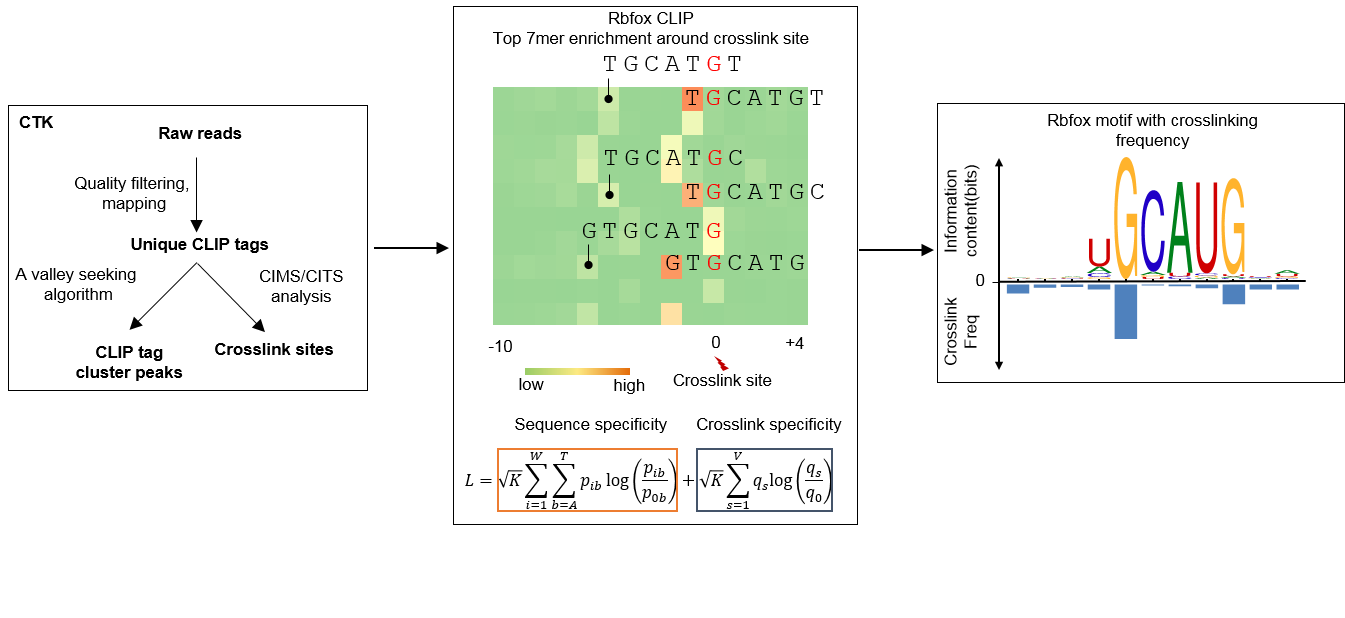
We applied mCross to a published CLIP datasets of a large panel of RBPs, including efficient CLIP (eCLIP) data of 112 unique RBPs from ENCODE. mCrossBase provides an interactive web interface to allow the research community to have easy access to this resource.