Research
From Zhang Laboratory
We are working on RNA at the interface of Systems Biology, Computer Science and Molecular Neuroscience, currently sponsored by NIH, Simons Foundation, and Columbia University Medical Center.
Overview
|
Post-transcriptional regulation at the RNA level has profound impact on gene expression, especial for the development and function of the nervous system. Such regulation is dictated by interaction of at least several hundred RNA-binding proteins (RBPs) with their target transcripts, or RNA-regulatory networks. Our current work focus on neuron-specific alternative splicing, one critical step of RNA regulation. Our research interests range from basic understanding of the specificity of protein-RNA interaction, to organization of splicing regulatory networks, and to function of specific splice variants.
|
Protein-RNA interactions at single nucleotide resolution
|
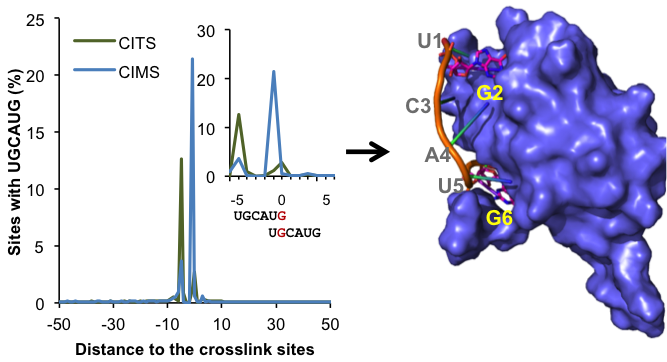
The RBP binding sites are notoriously difficult to map probably because of the flexibility of RNA. The development of cross linking and immunoprecipitation followed by high-throughput sequencing (HITS-CLIP or CLIP-Seq) has been a revolution to the RNA field. This high-throughput biochemical assay makes it possible to map in vivo protein-RNA interactions on a genome wide scale. Standard analysis pipelines of CLIP data typically map protein-RNA interaction sites at a resolution of ~30-60 nt. We developed a new computational method to determine the exact protein-RNA crosslink sites. This method, termed CIMS and CITS analysis, allows us to map protein-RNA interactions at single nucleotide resolution. We are using this powerful approach to obtain unprecedented details about how the specificity of protein-RNA interaction is determined.
|
Organizational principles and functional impact of neuronal RNA-regulatory networks
|
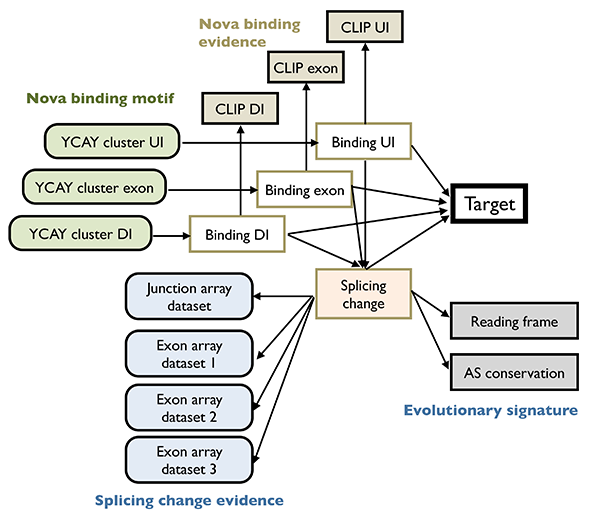
To understand the function of RNA regulatory network, a first step is to infer the structure of such networks, which is challenging due in part to the degeneracy and dynamic nature of the splicing code. We pioneered the development of integrative analysis of splicing regulatory networks, which has enabled us to identify alternative exons regulated by specific RBPs with unprecedented accuracy and sensitivity. In brief, our strategy was to identify altered splicing upon genetic RBP depletion, although such changes can be either direct or indirect. The latter was distinguished by mapping the precise RBP binding sites in vivo using HITS-CLIP. These data were then formally combined using a Bayesian network to determine high-confidence, direct target transcripts25,28. Studies using this paradigm demonstrated the concerted regulation of hundreds of alternative exons by individual neuronal RBPs. Investigation of the resulting networks allow us to make unexpected findings such as coupling of splicing with post-translational modifications.
|
RNA regulatory networks in neural development
|
For our current work, one focus is to investigate systematically how multiple RBPs work together to drive dynamic regulation during neurodevelopment. We aim to find predictive rules of combinatorial regulation, and how groups of RBPs define subsets of target transcripts that are functional distinct. To this end, we rely on the structure of RBPs defined by integrative modeling approaches, and statistical inference to determine the correlation structure underlying time series transcriptome data. Another exciting direction is the use of embryonic stem cells (ESCs) and in vitro differentiation of neurons from ESCs. We specifically focus on in vitro differentiation of motor neurons, a group of nerve cells through which the brain controls muscle contraction. Motor neurons are also the primary targets in several devastating neuron degenerative diseases including amyotrophic lateral sclerosis (ALS) and spinal muscular satrapy (SMA). This system has two key advantages: 1) the differential process recapitulate many key aspects of in vivo motor neuron development; 2) the use of ES cells makes it possible to leverage genome engineering techniques to perturb RNA regulatory networks. By dissecting RNA regulatory networks in detail, we hope to obtain a deeper understanding of their functional impact and the underlying regulatory mechanisms in this well defined, clinically relevant cell type. |
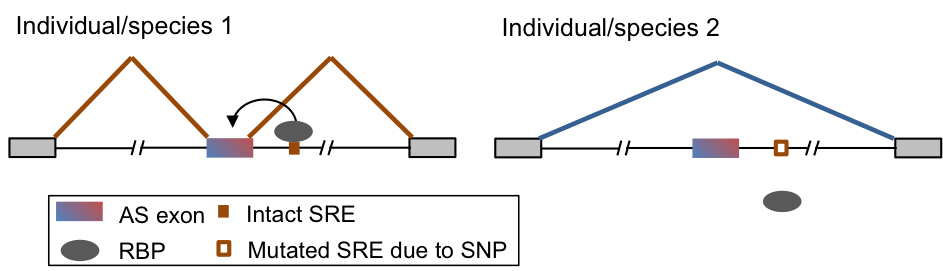
Variation of RNA-regulatory networks in evolution, human populations and in neuronal disorders
|
Another related direction of our lab is to evaluate the impact of mutations on RNA regulation in normal physiology or disease. Our study spans three contexts: comparison of different species (e.g. rodents and primates), different human populations, and patients affected by neurological diseases and normal controls. Such study is facilitated by our ability to determine protein-RNA interactions at a high resolution and distinction of functional vs. nonfunctional interactions. We are applying this strategy to parallel systems in different species that are directly comparable, large transcriptome profiles of human populations generated by consortium efforts, and mutations identified by genomic sequencing compiled from the public domain and collaborators.
|
High-throughput transcriptomic data analysis
|
Our work heavily relies on high-throughput technologies which produce enormous amount of data, and on algorithms to transform these data into useful information. We are interested in developing better algorithms to process transcroptomic data, such as mapping RNA-Seq reads, discovering and quantifying RNA processing in specific conditions, and modeling the specificity of protein-RNA interactions.
|