Difference between revisions of "Main Page"
From Zhang Laboratory
(→Lab News) |
|||
| (369 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{#meta: keywords | computational biology; systems biology; RNA splicing and regulatory networks; gene expression }}{{#meta: description | The Chaolin Zhang Laboratory Home Page at Columbia University}}{{#meta: Content-Type | text/html; charset=utf-8 }} | {{#meta: keywords | computational biology; systems biology; RNA splicing and regulatory networks; gene expression }}{{#meta: description | The Chaolin Zhang Laboratory Home Page at Columbia University}}{{#meta: Content-Type | text/html; charset=utf-8 }} | ||
| + | <div style='text-align: right;'> | ||
| + | [[File:Twitter.jpg|25px|link=https://twitter.com/chaolinzhang]] [[File:LinkedIn.png|20px|link=https://www.linkedin.com/in/chaolin-zhang-52835217/]] | ||
| + | </div> | ||
<html> | <html> | ||
| − | <div | + | |
| − | <img src="/ | + | <script type="text/javascript" src="/data/Jssor/js/Jssor.Core.js"></script> |
| − | </div> | + | <script type="text/javascript" src="/data/Jssor/js/Jssor.Debug.js"></script> |
| + | <script type="text/javascript" src="/data/Jssor/js/Jssor.EventManager.js"></script> | ||
| + | <script type="text/javascript" src="/data/Jssor/js/Jssor.Easing.js"></script> | ||
| + | <script type="text/javascript" src="/data/Jssor/js/Jssor.Point.js"></script> | ||
| + | <script type="text/javascript" src="/data/Jssor/js/Jssor.Utils.js"></script> | ||
| + | <script type="text/javascript" src="/data/Jssor/js/Jssor.Navigator.js"></script> | ||
| + | <script type="text/javascript" src="/data/Jssor/js/Jssor.CaptionSlider.js"></script> | ||
| + | <script type="text/javascript" src="/data/Jssor/js/Jssor.Slider.js"></script> | ||
| + | <script type="text/javascript" src="/data/Jssor/js/Jssor.ThumbnailNavigator.js"></script> | ||
| + | <script> | ||
| + | jssor_slider1_starter = function (containerId) { | ||
| + | var _CaptionTransitions = []; | ||
| + | //Left to Right | ||
| + | _CaptionTransitions["L-R"] = { $Duration: 600, $FlyDirection: 1 }; | ||
| + | //Right to Left | ||
| + | _CaptionTransitions["R-L"] = { $Duration: 600, $FlyDirection: 2 }; | ||
| + | //Top to Bottom | ||
| + | _CaptionTransitions["T-B"] = { $Duration: 600, $FlyDirection: 4 }; | ||
| + | //Bottom to Top | ||
| + | _CaptionTransitions["B-T"] = { $Duration: 600, $FlyDirection: 8 }; | ||
| + | |||
| + | //To build caption transition code, reference to http://slideshow.jssor.com/documentation/transition-builder-caption.html | ||
| + | |||
| + | var jssor_slider1 = new $JssorSlider$(containerId, { | ||
| + | $SlideDuration:1200, | ||
| + | $HoverToPause: true, | ||
| + | $ShowLoading: true, //[Optional] Show loading screen or not default value is false | ||
| + | $AutoPlay: true, //[Optional] Whether to auto play, default value is false | ||
| + | $PlayOrientation: 1, //[Optional] Orientation to play slide (for auto play, navigation), 1 horizental, 2 vertical, default value is 1 | ||
| + | $DragOrientation: 3, //[Optional] Orientation to drag slide, 0 no drag, 1 horizental, 2 vertical, 3 either, default value is 1 (Note that the $DragOrientation should be the same as $PlayOrientation when $DisplayPieces is greater than 1, or parking position is not 0) | ||
| + | |||
| + | $CaptionSliderOptions: { //[Optional] Options which specifies how to animate caption | ||
| + | $Class: $JssorCaptionSlider$, //[Required] Class to create instance to animate caption | ||
| + | $CaptionTransitions: _CaptionTransitions, //[Required] Caption transitions to play caption, see caption transition section at jssor slideshow transition builder | ||
| + | $PlayInMode: 1, //[Optional] 0 None (no play), 1 Chain (goes after main slide), 2 Parallel (goes synchronous with main slide), default value is 1 | ||
| + | $PlayOutMode: 1 //[Optional] 0 None (no play), 1 Chain (goes before main slide), 2 Parallel (goes synchronous with main slide), default value is 1 | ||
| + | } | ||
| + | }); | ||
| + | } | ||
| + | </script> | ||
| + | |||
| + | <center> | ||
| + | <div id="slider1_container" class="slider1" style="position: relative; width: 600px; height: 300px;"> | ||
| + | |||
| + | <!-- Loading Screen --> | ||
| + | <div u="loading" style="position: absolute; top: 0px; left: 0px;"> | ||
| + | <div style="filter: alpha(opacity=70); opacity:0.7; position: absolute; display: block; | ||
| + | background-color: #000000; top: 0px; left: 0px;width: 100%;height:100%;"> | ||
| + | </div> | ||
| + | <div style="position: absolute; display: block; background: url(/data/Jssor/img/loading.gif) no-repeat center center; | ||
| + | top: 0px; left: 0px;width: 100%;height:100%;"> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <!-- Slides Container --> | ||
| + | <div u="slides" style="position: absolute; left: 0px; top: 0px; width:600px; height:300px; overflow: hidden;"> | ||
| + | |||
| + | <div> | ||
| + | <a u=image href="/index.php/Research" title="Our long term goal: better mechanistic understanding of RNA splicing regulation and function." rel="nofollow"><img u="image" src="/data/images/slideshow/deconstruct_splice_code.png" width="600" height="300" /></a> | ||
| + | </div> | ||
| + | |||
| + | <div> | ||
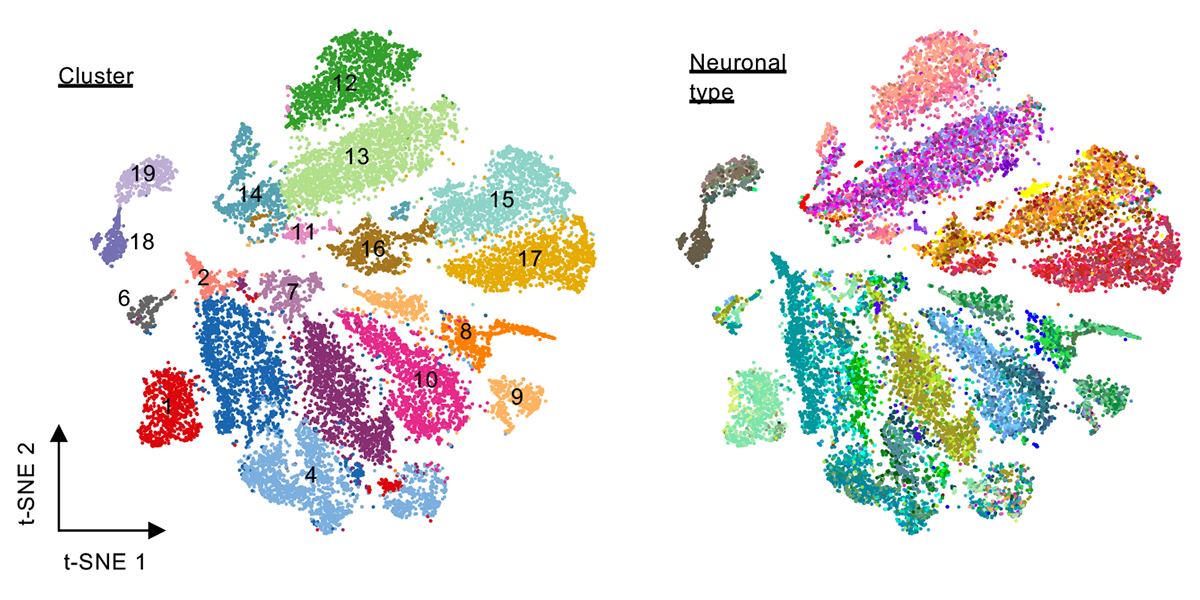
| + | <a u=image href="https://www.pnas.org/content/118/10/e2013056118/tab-article-info" title="Clustering of neuronal subtypes based on splicing profiles (PNAS, 2021)." rel="nofollow"><img u="image" src="/data/images/slideshow/neuronal_subtype_splicing.png" width="600" height="300" /></a> | ||
| + | </div> | ||
| + | |||
| + | <div> | ||
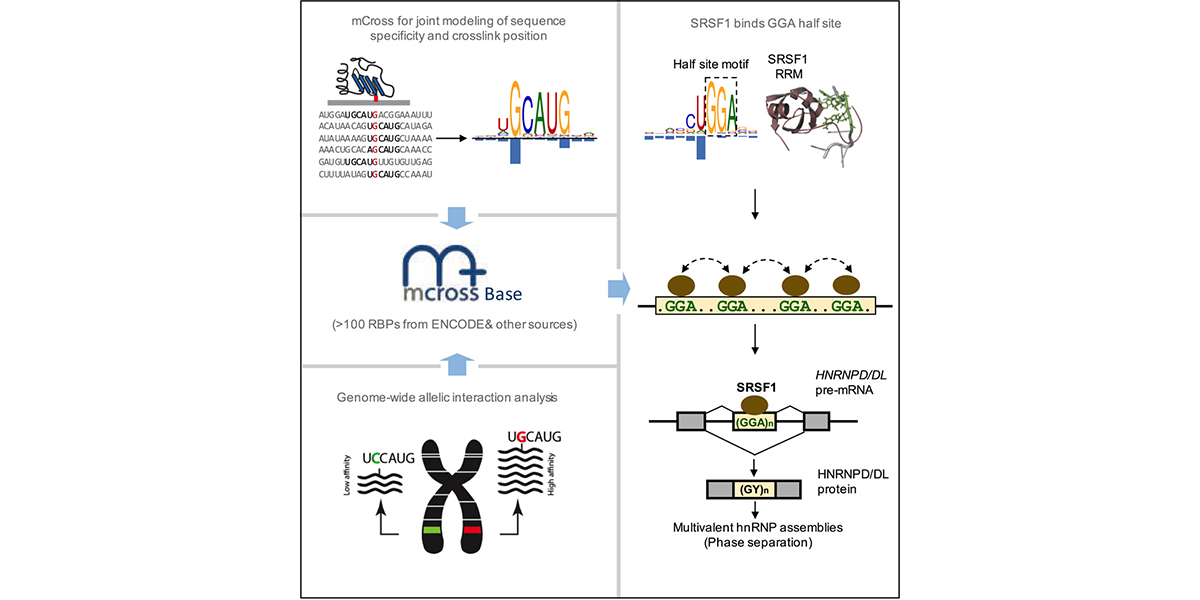
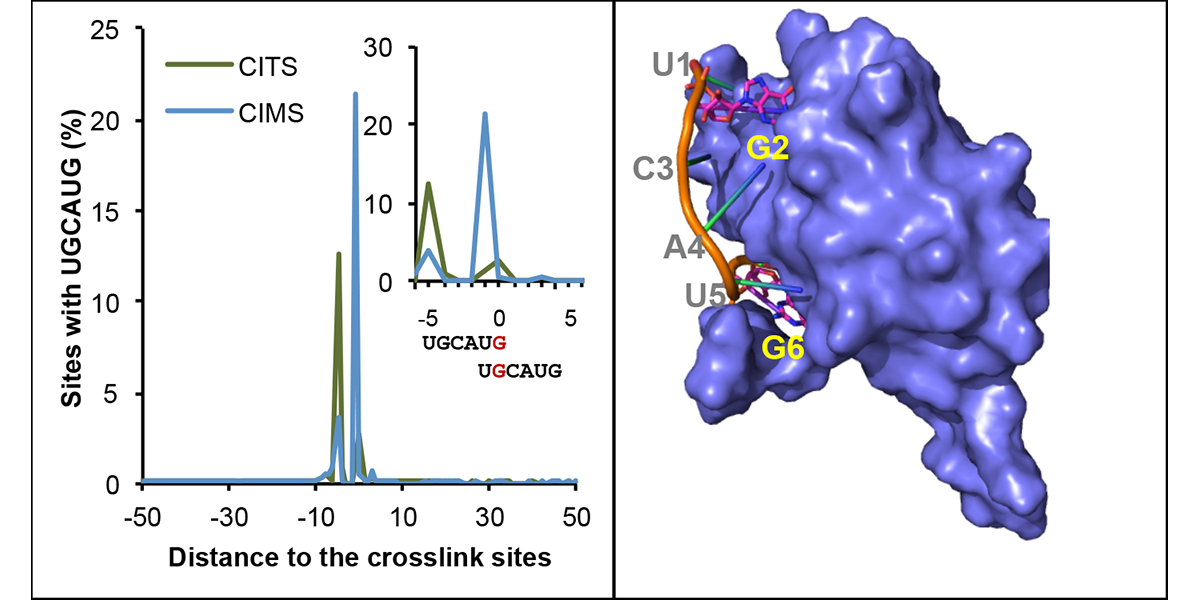
| + | <a u=image href="https://www.sciencedirect.com/science/article/pii/S1097276519300929" title="mCross: precise registration of protein-RNA crosslink sites to define RBP binding specificity using CLIP data (Mol. Cell, 2019)." rel="nofollow"><img u="image" src="/data/images/slideshow/mcross.png" width="600" height="300" /></a> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div> | ||
| + | <a u=image href="https://authors.elsevier.com/a/1XPn83vVUP60qL" title="A subclass of let-7 microRNA partially escape from LIN28 repression (Mol. Cell, 2018)." rel="nofollow"><img u="image" src="/data/images/slideshow/lin28.png" width="600" height="300" /></a> | ||
| + | </div> | ||
| + | |||
| + | <div> | ||
| + | <a u=image href="https://www.nature.com/articles/s41467-018-04559-0" title="Precise timing of alternative splicing switches during neuronal development (Nat. Commun., 2018)" rel="nofollow"><img u="image" src="/data/images/slideshow/devas.png" width="600" height="300" /></a> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div> | ||
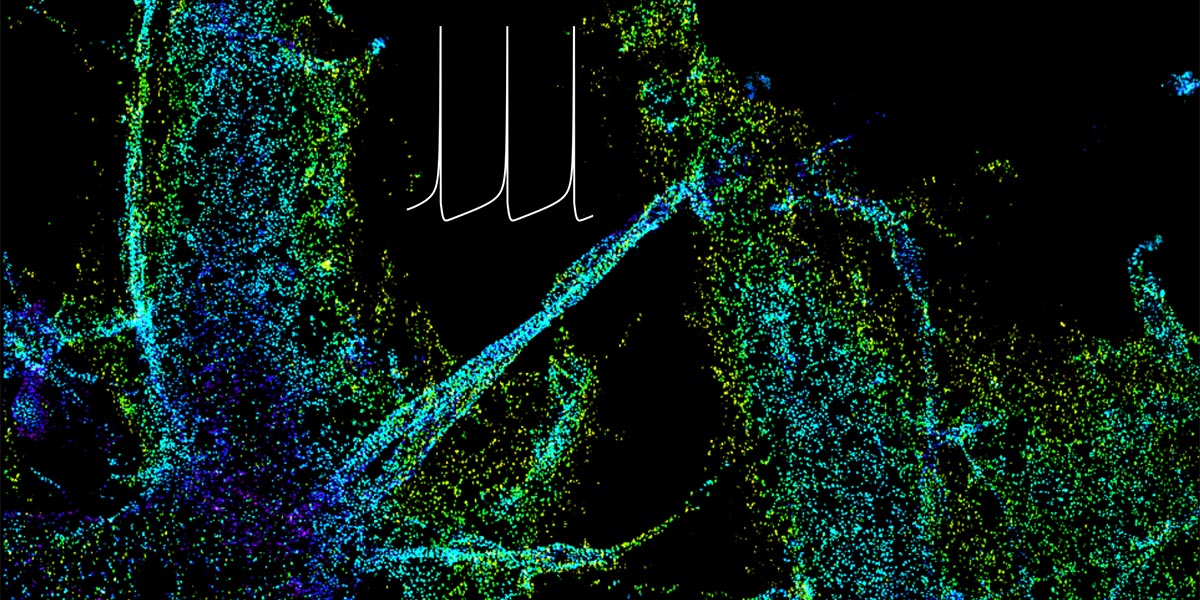
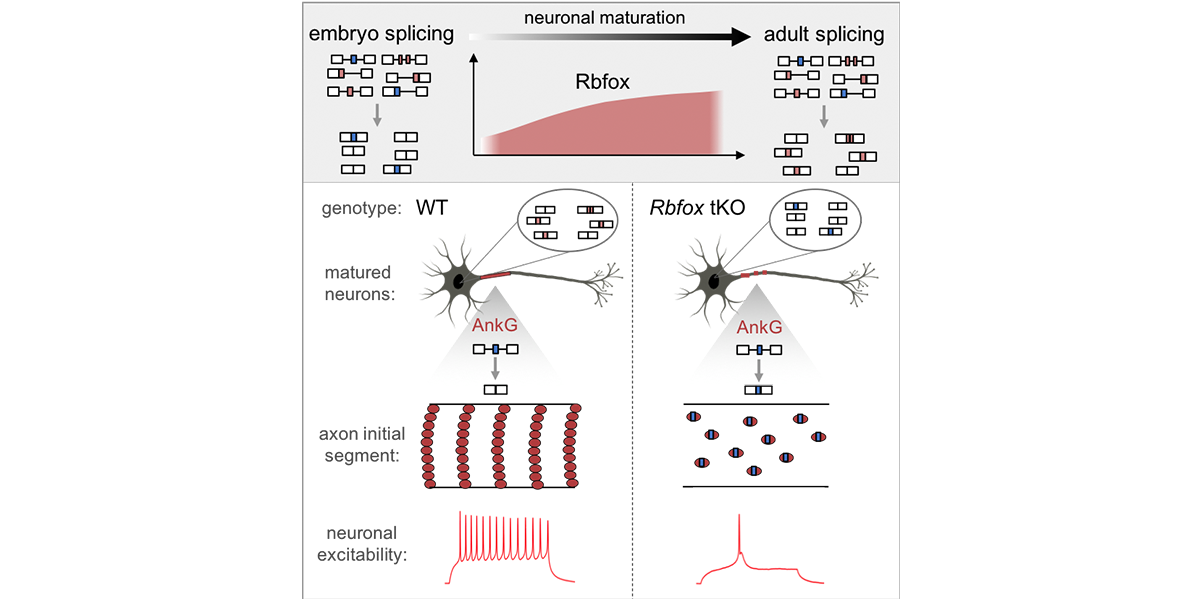
| + | <a u=image href="http://www.cell.com/neuron/fulltext/S0896-6273(18)30023-0" title="Rbfox regulates axon initial segment and neuronal excitability (Neuron, 2018)." rel="nofollow"><img u="image" src="/data/images/slideshow/ais.jpg" width="600" height="300" /></a> | ||
| + | </div> | ||
| + | |||
| + | <div> | ||
| + | <a u=image href="http://www.cell.com/neuron/fulltext/S0896-6273(18)30023-0" title="Rbfox regulates axon initial segment and neuronal excitability (Neuron, 2018)." rel="nofollow"><img u="image" src="/data/images/slideshow/RbfoxMN.png" width="600" height="300" /></a> | ||
| + | </div> | ||
| + | |||
| + | <div> | ||
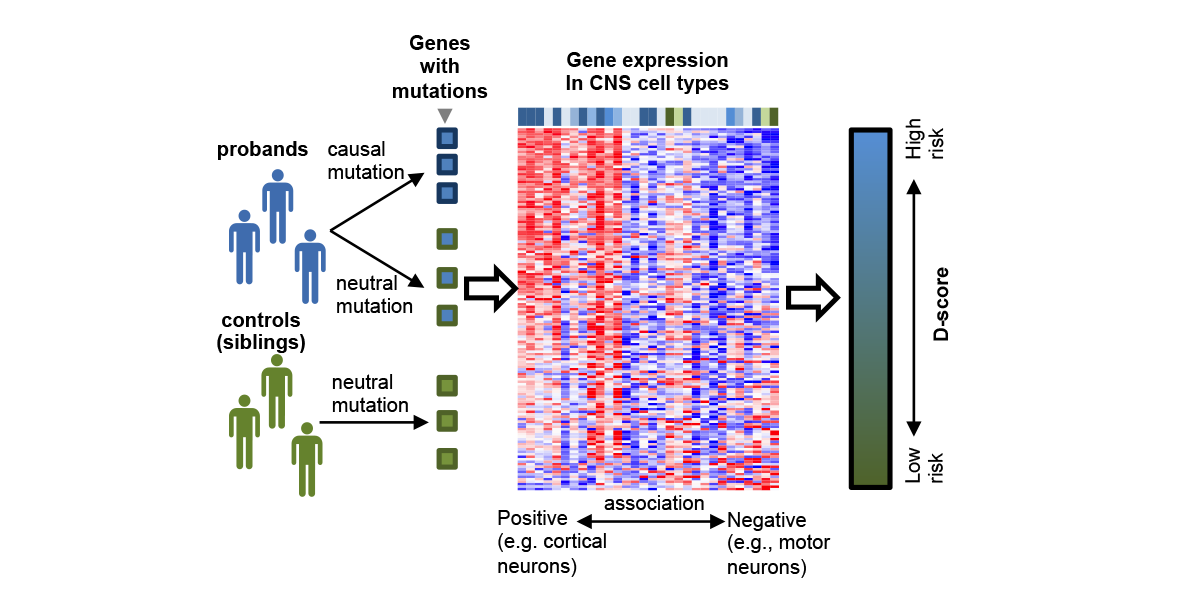
| + | <a u=image href="http://onlinelibrary.wiley.com/doi/10.1002/humu.23147/abstract" title="A neuronal-cell type-specific gene expression signature predicts autism risk genes (Hum. Mut. 2017)." rel="nofollow"><img u="image" src="/data/images/slideshow/DAMAGES.png" width="600" height="300" /></a> | ||
| + | </div> | ||
| + | |||
| + | <div> | ||
| + | <a u=image href="/index.php/Research" title="Genes have 5' splice sites and 3' splice sites, but some splice sites have dual roles" rel="nofollow"><img u="image" src="/data/images/slideshow/dual_splice_site.png" width="600" height="300" /></a> | ||
| + | </div> | ||
| + | |||
| + | <div> | ||
| + | <a u=image href="/index.php/Research" title="UV-crosslinking induced protein-RNA adducts provide a signature to determine protein-RNA interactions at single nucleotide resolution" rel="nofollow"><img u="image" src="/data/images/slideshow/crosslink.png" width="600" height="300" /></a> | ||
| + | </div> | ||
| + | |||
| + | <div> | ||
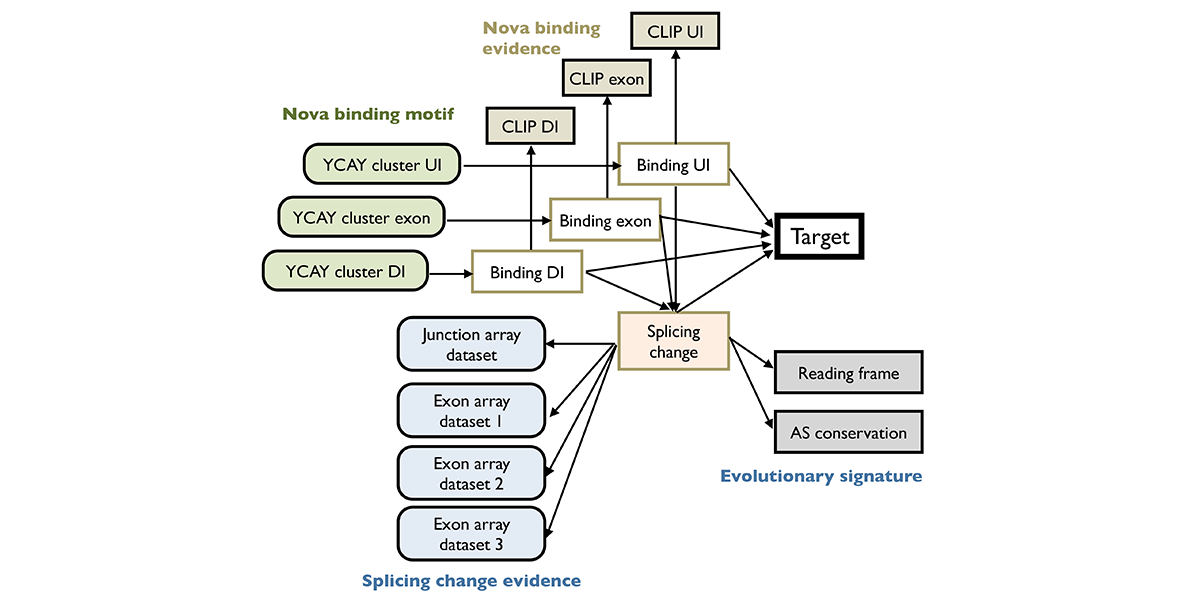
| + | <a u=image href="/index.php/Research" title="An integrative modeling approach to construct splicing regulatory networks" rel="nofollow"><img u="image" src="/data/images/slideshow/bnet.png" width="600" height="300" /></a> | ||
| + | </div> | ||
| + | |||
| + | <div> | ||
| + | <a u=image href="/index.php/Research" title="We aim to understand the contribution to alternative splicing to transcriptome diversity in brain development" rel="nofollow"><img u="image" src="/data/images/slideshow/brain_dev.png" width="600" height="300" /></a> | ||
| + | </div> | ||
| + | |||
| + | <div> | ||
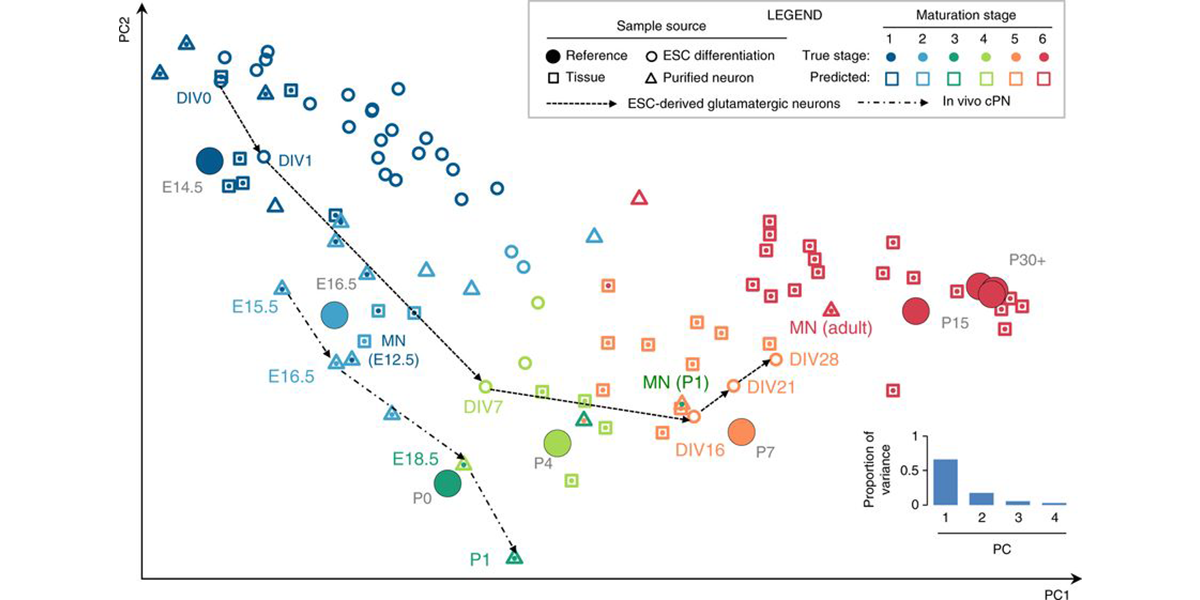
| + | <a u=image href="/index.php/Research" title="ESC-derived motor neuronsprovide an ideal system to model neural development and maturation in vitro" rel="nofollow"><img u="image" src="/data/images/slideshow/eb.png" width="600" height="300" /></a> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | <a style="display:none" href="http://slideshow.jssor.com">Javascript Slideshow</a> | ||
| + | <!-- Trigger --> | ||
| + | <script> | ||
| + | jssor_slider1_starter('slider1_container'); | ||
| + | </script> | ||
| + | </div> | ||
| + | |||
| + | </center> | ||
| + | |||
| + | |||
| + | |||
</html> | </html> | ||
| − | |||
| − | + | ==Introduction of the Zhang Laboratory== | |
| + | {|class="zebra wikitable" width="100%" style="border:1px solid; border-collapse:collapse;" | ||
| − | + | |- | |
| + | |style="width: 1px"| | ||
| − | We | + | We are part of the [http://systemsbiology.columbia.edu Department of Systems Biology], the [http://cpmcnet.columbia.edu/dept/gsas/biochem/ Department of Biochemistry and Molecular Biophysics], and the [http://www.columbiamnc.org/ Motor Neuron Center] at [http://http://www.cumc.columbia.edu Columbia University Medical Center]. |
| + | We are fascinated by the complexity of the mammalian nervous system and its underlying molecular mechanisms. While mammals have a similar number of genes compared to phenotypically simpler organisms (such as worms), one apparent feature of mammalian genes is their more complicated gene structures, providing an opportunity for sophisticated regulation at the RNA level. | ||
| + | |||
| + | The focus of the Zhang lab is to dissect RNA regulatory networks in the nervous system as a way to understand the mammalian complexity manifested in evolutionary-developmental (evo-devo) processes and in several neuronal disorders. The lab has a mixed dry and wet setup, and takes a multidisciplinary approach that tightly integrates high-throughput biochemistry, genomics and computational approaches applied to mouse models and in vitro neuronal differentiation systems from pluripotent stem cells. On the mechanistic side, the Zhang lab focuses on fundamental understanding of the targeting specificity of RNA-binding proteins (RBPs), how they regulate alternative splicing in various cellular contexts, especially in the nervous system, and how such regulation can be disrupted by mutations and genetic variations. On the functional side, the lab aims to uncover the roles of RBPs in determining the neuronal cell fate, morphological and functional properties during neural differentiation and maturation. The lab has also been working on translating fundamental knowledge on RNA regulation to precision genetic medicine, with a particular focus on multiple devastating monogenic diseases affecting the central nervous system. | ||
| + | |||
| + | |||
| + | [[File:Lab photo 2024 v3s.jpg|720px|center|link=]] | ||
| + | |||
| + | <div style="text-align:center"> | ||
| + | 2024 Lab Photo - Celebrating Summer Undergrads Amanda Lopez-Ramirez and Chamari Abercrombie, first-row middle and right. | ||
| + | |||
| + | (Chamari will stay as a post-bac for the next two years) | ||
| + | </div> | ||
| + | |} | ||
| + | |||
| + | ==Lab News== | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |style="width: 50%;vertical-align:top| | ||
| + | |||
| + | [[File:Terrarium2024.jpg|600px|link=http://www.facebook.com/media/set/?set=a.882018307363416&type=3]] | ||
| + | |||
| + | Making self-sustaining terrariums (Oct, 2024) | ||
| + | |||
| + | |style="width: 50%;vertical-align:top| | ||
| + | |||
| + | * 09/25/2024. Congrats Ruchika for winning the best poster award during the annual department retreat! | ||
| + | * 09/16/2024. Welcome Cynthia to rotate in the Zhang lab. Cynthia will join an exciting project to develop tools for programmable RNA targeting! | ||
| + | * 08/09/2024. Our DeltaSplice paper is officially out in [http://genome.cshlp.org/content/early/2024/08/05/gr.279044.124 Genome Research]. | ||
| + | * 07/08/2024. Welcome Amanda Lopez-Ramirez ([http://www.gsas.columbia.edu/content/summer-research-program SRP program]), Camari Abercrombie ([http://www.neurosciencephd.columbia.edu/content/CADRE-postbac-program CADRE program]), and Bryan Shi for joining us in the summer. Camari will actually work with us as a post-bac in the next two years! | ||
| + | * 07/08/2024. Welcome Tianji Yu, who officially joined the Zhang lab - Tianji is our 5th graduate student! | ||
| + | * 06/27/2024. Chaolin joined Harris Wang, interim department chair, to attend the "Meet the Faculty" panel discussion with DSB trainees. | ||
| + | * 06/26/2024. We are one of the four teams winning the 2024 [http://www.precisionmedicine.columbia.edu/content/funding-opportunities Columbia Precision Medicine Joint Pilot grant]. Thank Roy and Diana Vagelos for the generous support! | ||
| + | * 06/21/2024. Preprint release - Dan Moakley (recent graduate)'s PhD work to infer neuron type-specific splicing-regulatory networks from single-cell RNA-seq transcriptome: http://doi.org/10.1101/2024.06.13.597128. | ||
| + | |||
| + | |||
| + | |||
| + | <div style="text-align:right"> | ||
| + | >>> [[Lab news|all lab news]] | ||
| + | </div> | ||
| + | |} | ||
| + | |||
| + | ==Where We Are== | ||
| + | |||
| + | {|class="zebra wikitable" width="100%" style="border:1px solid; border-collapse:collapse;" | ||
| + | |||
| + | |- | ||
| + | |style="width: 1px" align="center"| | ||
| + | |||
| + | [[File:Cumc2.jpg|600px|Columbia University Medical Center is located in the Washington Heights neighborhood of Manhattan in New York City. It is situated at the northern tip of Manhattan, at the intersection of West 168th Street and Broadway. The Medical Center is easily accessible by public transportation, with the 1, A and C trains stopping at the 168th Street station.|link=]] | ||
| + | |||
| + | | | ||
| + | |||
| + | [[File:Washingtonheight.jpeg|320px|link=]] | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | ||
| + | <div style="text-align:center"> | ||
| + | Columbia University Irving Medical Center (image credit: [http://www.pinterest.com/pin/columbia-university-medical-center-partially-manhatten-nyc--619385754984816311/ internet]) | ||
| + | </div> | ||
| + | | | ||
| + | <div style="text-align:center"> | ||
| + | Washington Height, NYC (image credit: [http://twitter.com/KellyrKopp @KellyrKopp/twitter]) | ||
| + | </div> | ||
| + | |} | ||
<html> | <html> | ||
| + | |||
<div align="center"> | <div align="center"> | ||
| − | <a href="http:// | + | |
| − | <a href="http://www.columbiamnc.org/"><img src=" | + | |
| − | <a href="http://www.c2b2.columbia.edu"><img src=" | + | <a href="http://systemsbiology.columbia.edu"><img src="/data/images/CSB.png" width="147" height="40" /></a> |
| + | <a href="http://www.biochem.cuimc.columbia.edu"><img src="/images/d/d6/Logo_biochem.jpg" width="206" height="40" /></a> | ||
| + | <a href="http://www.columbiamnc.org/"><img src="/data/images/mnc_logo.gif" width="122" height="35" /></a> | ||
| + | <a href="http://www.c2b2.columbia.edu"><img src="/data/images/C2B2_logo.png" width="215" height="60" /></a> <p> | ||
| + | |||
</div> | </div> | ||
| + | |||
| + | |||
</html> | </html> | ||
Latest revision as of 15:23, 17 October 2024
Introduction of the Zhang Laboratory
|
We are part of the Department of Systems Biology, the Department of Biochemistry and Molecular Biophysics, and the Motor Neuron Center at Columbia University Medical Center. We are fascinated by the complexity of the mammalian nervous system and its underlying molecular mechanisms. While mammals have a similar number of genes compared to phenotypically simpler organisms (such as worms), one apparent feature of mammalian genes is their more complicated gene structures, providing an opportunity for sophisticated regulation at the RNA level. The focus of the Zhang lab is to dissect RNA regulatory networks in the nervous system as a way to understand the mammalian complexity manifested in evolutionary-developmental (evo-devo) processes and in several neuronal disorders. The lab has a mixed dry and wet setup, and takes a multidisciplinary approach that tightly integrates high-throughput biochemistry, genomics and computational approaches applied to mouse models and in vitro neuronal differentiation systems from pluripotent stem cells. On the mechanistic side, the Zhang lab focuses on fundamental understanding of the targeting specificity of RNA-binding proteins (RBPs), how they regulate alternative splicing in various cellular contexts, especially in the nervous system, and how such regulation can be disrupted by mutations and genetic variations. On the functional side, the lab aims to uncover the roles of RBPs in determining the neuronal cell fate, morphological and functional properties during neural differentiation and maturation. The lab has also been working on translating fundamental knowledge on RNA regulation to precision genetic medicine, with a particular focus on multiple devastating monogenic diseases affecting the central nervous system.
 2024 Lab Photo - Celebrating Summer Undergrads Amanda Lopez-Ramirez and Chamari Abercrombie, first-row middle and right. (Chamari will stay as a post-bac for the next two years) |
Lab News
|
Making self-sustaining terrariums (Oct, 2024) |
>>> all lab news |
Where We Are
|
|
|
|
Columbia University Irving Medical Center (image credit: internet) |
Washington Height, NYC (image credit: @KellyrKopp/twitter) |