Difference between revisions of "Main Page"
From Zhang Laboratory
| Line 168: | Line 168: | ||
|style="width: 50%;vertical-align:top| | |style="width: 50%;vertical-align:top| | ||
| + | * 10/17/2023. Congrats Yocelyn Recinos for winning the poster award at Systems Biology department retreat! | ||
* 10/02/2023. (Belated news) We released our [http://www.biorxiv.org/content/10.1101/2023.08.21.554109v1 preprint] on SpliceRUSH, a high-throughput screening method for mapping both proximal and distal splicing-regulatory elements in native sequences. Congrats Yocelyn and team! | * 10/02/2023. (Belated news) We released our [http://www.biorxiv.org/content/10.1101/2023.08.21.554109v1 preprint] on SpliceRUSH, a high-throughput screening method for mapping both proximal and distal splicing-regulatory elements in native sequences. Congrats Yocelyn and team! | ||
* 10/02/2023. (Belated news) Welcome new PhD students Albertine Albertine Neal, Fahad Paryani, and Tianji Yu for rotating in the lab! | * 10/02/2023. (Belated news) Welcome new PhD students Albertine Albertine Neal, Fahad Paryani, and Tianji Yu for rotating in the lab! | ||
| Line 176: | Line 177: | ||
*05/10/2023. Congrats Yocelyn Recinos for her successful PhD thesis defense! | *05/10/2023. Congrats Yocelyn Recinos for her successful PhD thesis defense! | ||
* 01/23/2023. Our [http://systemsbiology.columbia.edu department] highlighted our work in the [http://systemsbiology.columbia.edu/newsletters annual news letter]. | * 01/23/2023. Our [http://systemsbiology.columbia.edu department] highlighted our work in the [http://systemsbiology.columbia.edu/newsletters annual news letter]. | ||
| − | |||
Revision as of 06:32, 17 October 2023
Introduction of the Zhang Laboratory
|
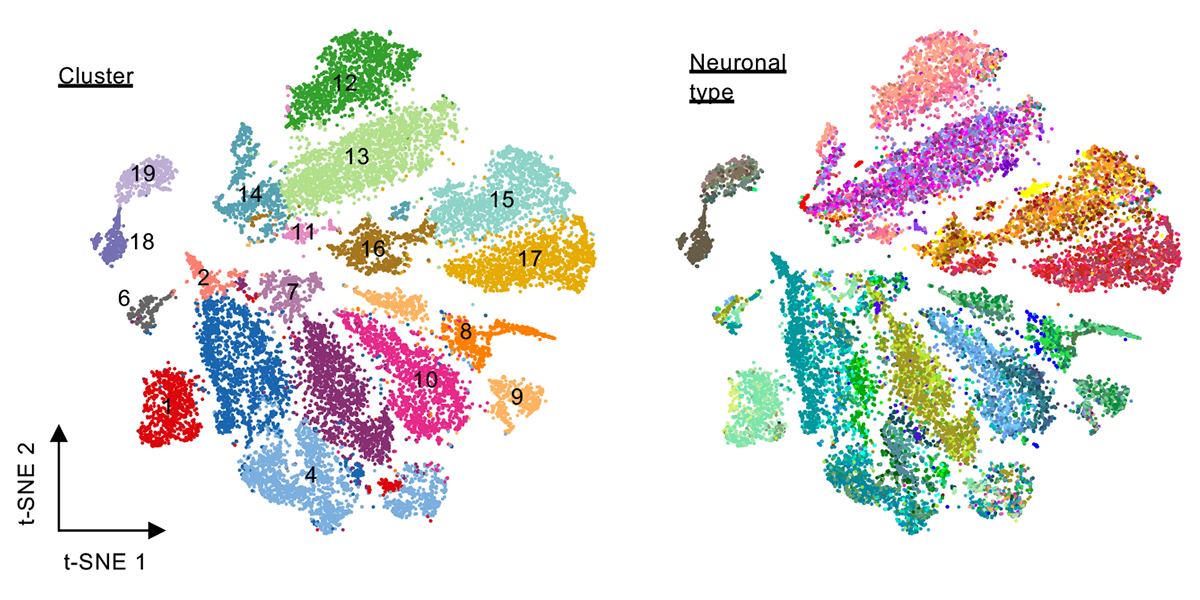
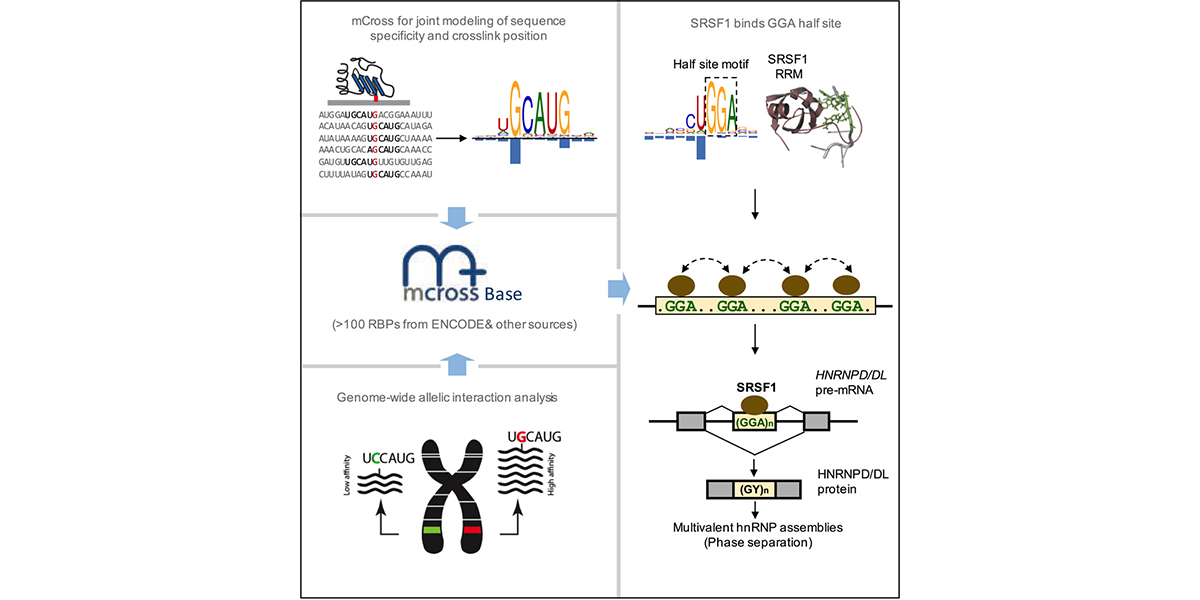
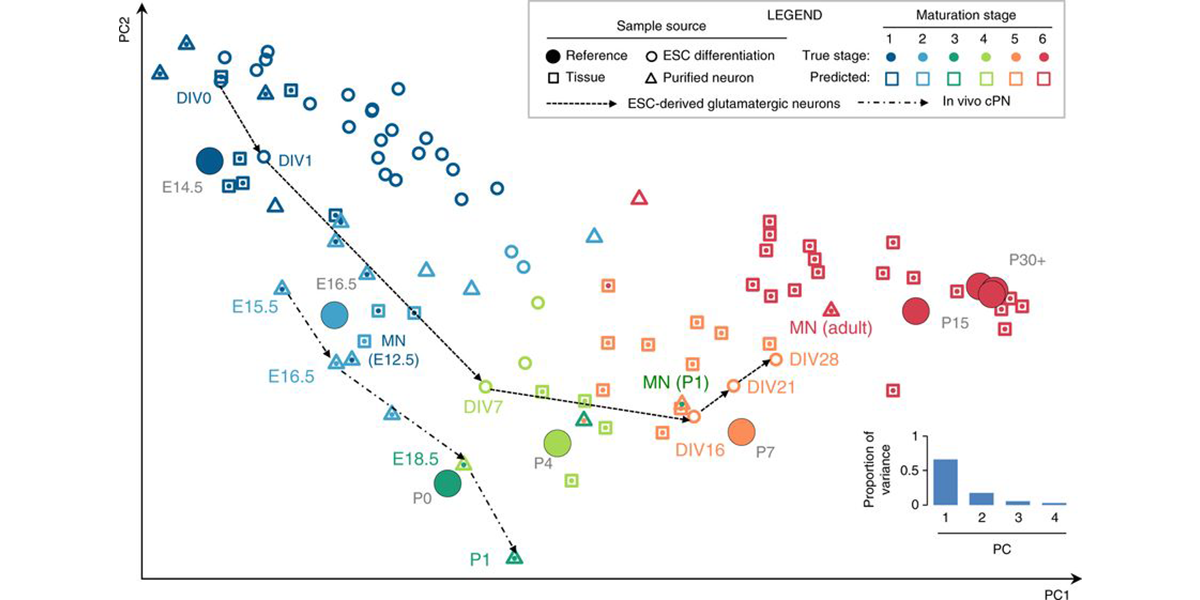
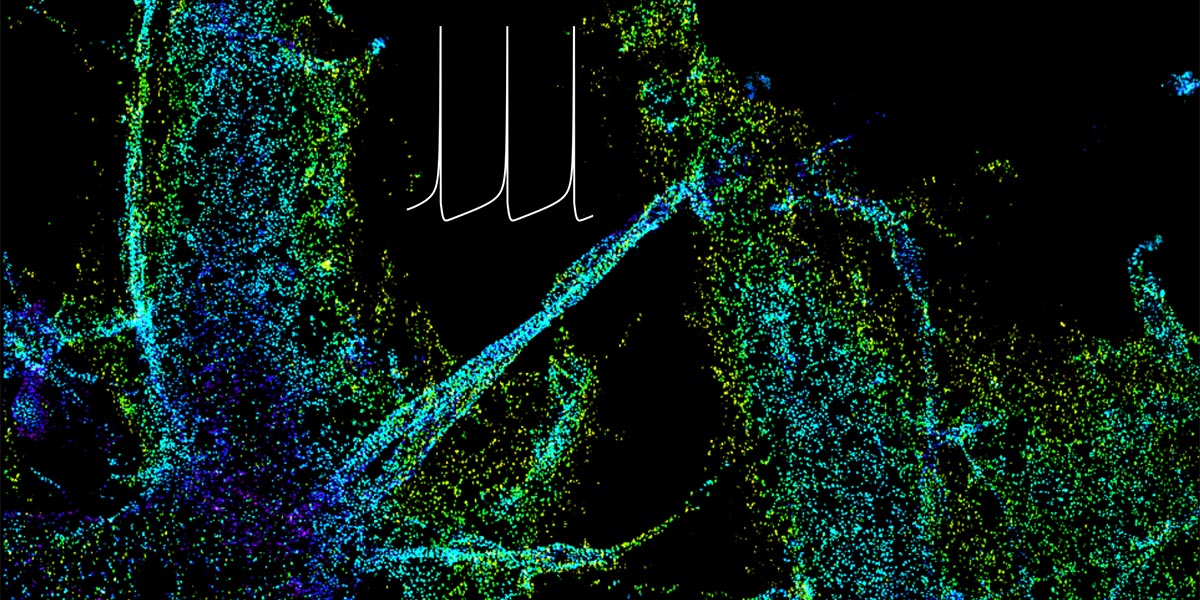
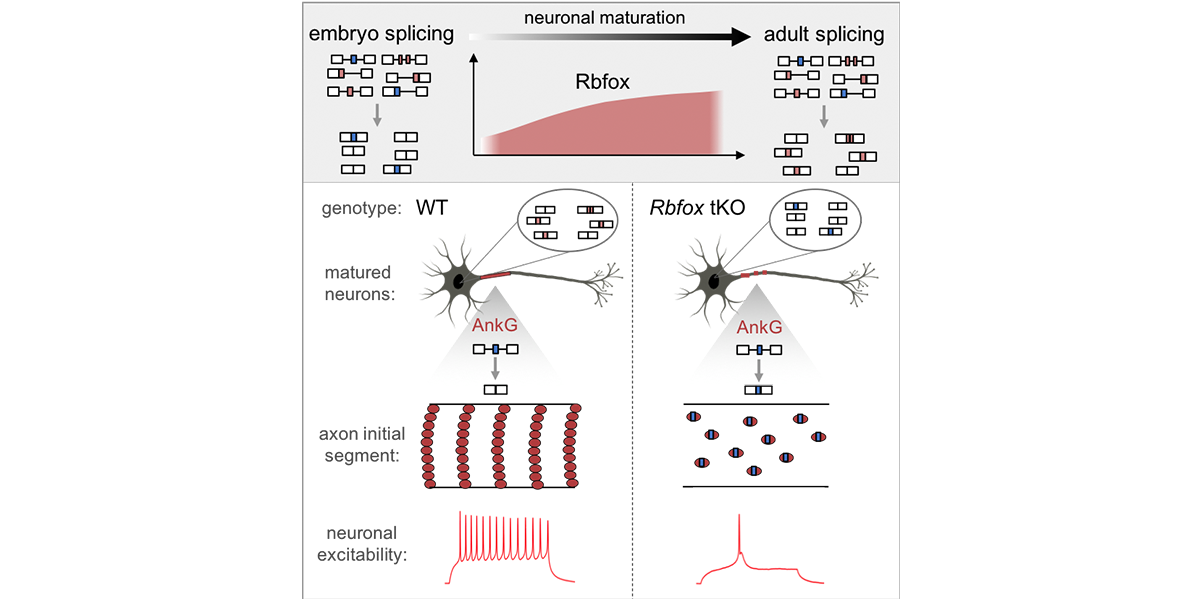
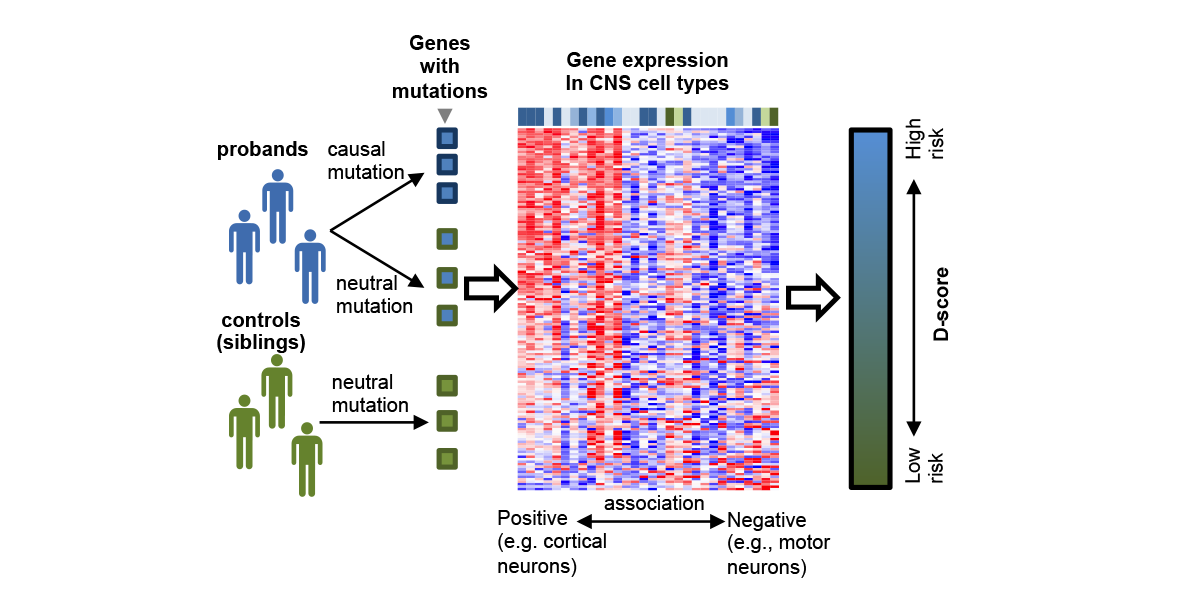
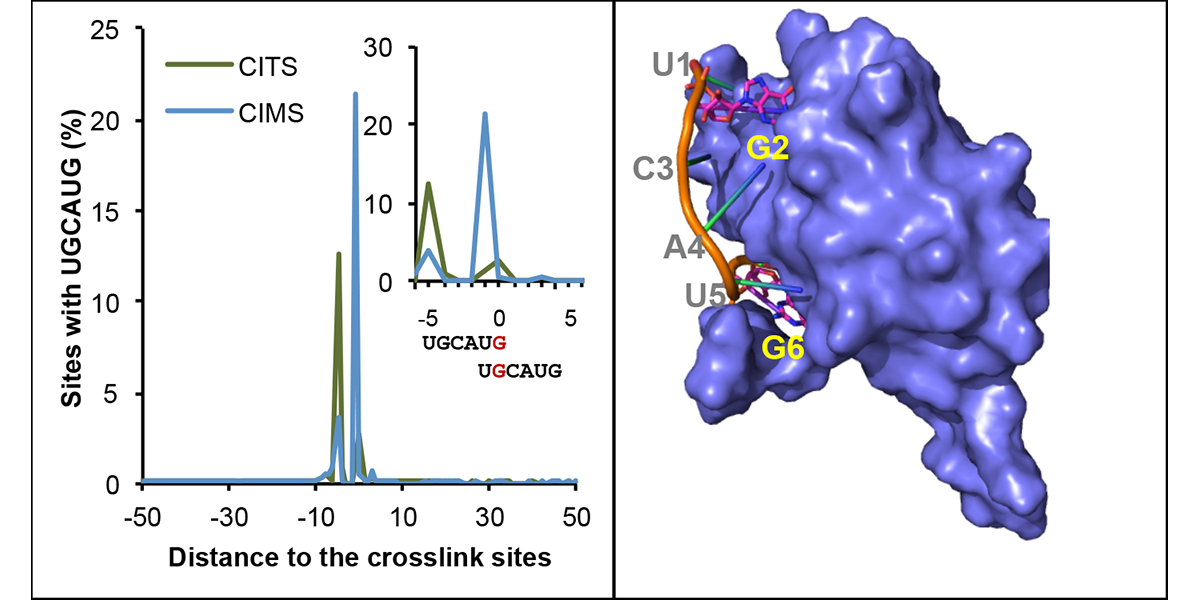
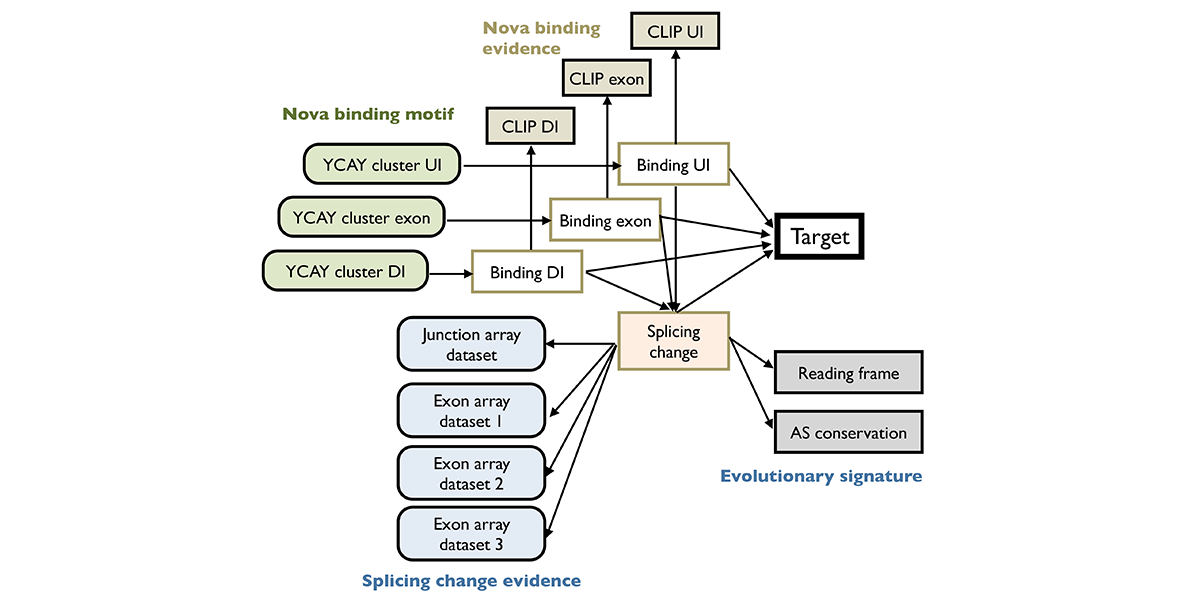
We are part of the Department of Systems Biology, the Department of Biochemistry and Molecular Biophysics, and the Motor Neuron Center at Columbia University Medical Center. We are fascinated by the complexity of the mammalian nervous system and its underlying molecular mechanisms. While mammals have a similar number of genes compared to phenotypically simpler organisms (such as worms), one apparent feature of mammalian genes is their more complicated gene structures, providing an opportunity for sophisticated regulation at the RNA level. The focus of the Zhang lab is to dissect RNA regulatory networks in the nervous system as a way to understand the mammalian complexity manifested in evolutionary-developmental (evo-devo) processes and in several neuronal disorders. The lab has a mixed dry and wet setup, and takes a multidisciplinary approach that tightly integrates high-throughput biochemistry, genomics and computational approaches applied to mouse models and in vitro neuronal differentiation systems from pluripotent stem cells. On the mechanistic side, the Zhang lab focuses on fundamental understanding of the targeting specificity of RNA-binding proteins (RBPs), how they regulate alternative splicing in various cellular contexts, especially in the nervous system, and how such regulation can be disrupted by mutations and genetic variations. On the functional side, the lab aims to uncover the roles of RBPs in determining the neuronal cell fate, morphological and functional properties during neural differentiation and maturation. The lab has also been working on translating fundamental knowledge on RNA regulation to precision genetic medicine, with a particular focus on multiple devastating monogenic diseases affecting the central nervous system.
 2022 Lab summer BBQ (10 year lab anniversary & celebration of Melissa's new faculty position) |
Lab News
|
Congrats, Dr. Recinos!! (05/2023) |
>>> all lab news |
Where We Are
|
|
|
|
Columbia University Irving Medical Center (image credit: internet) |
Washington Height, NYC (image credit: @KellyrKopp/twitter) |